The ER membrane protein complex restricts mitophagy by controlling BNIP3 turnover
- PMID: 38177312
- PMCID: PMC10883272
- DOI: 10.1038/s44318-023-00006-z
The ER membrane protein complex restricts mitophagy by controlling BNIP3 turnover
Abstract
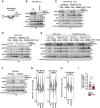
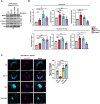
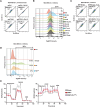
Lysosomal degradation of autophagy receptors is a common proxy for selective autophagy. However, we find that two established mitophagy receptors, BNIP3 and BNIP3L/NIX, are constitutively delivered to lysosomes in an autophagy-independent manner. This alternative lysosomal delivery of BNIP3 accounts for nearly all its lysosome-mediated degradation, even upon mitophagy induction. To identify how BNIP3, a tail-anchored protein in the outer mitochondrial membrane, is delivered to lysosomes, we performed a genome-wide CRISPR screen for factors influencing BNIP3 flux. This screen revealed both known modifiers of BNIP3 stability as well as a pronounced reliance on endolysosomal components, including the ER membrane protein complex (EMC). Importantly, the endolysosomal system and the ubiquitin-proteosome system regulated BNIP3 independently. Perturbation of either mechanism is sufficient to modulate BNIP3-associated mitophagy and affect underlying cellular physiology. More broadly, these findings extend recent models for tail-anchored protein quality control and install endosomal trafficking and lysosomal degradation in the canon of pathways that tightly regulate endogenous tail-anchored protein localization.
Keywords: BNIP3; EMC; Mitophagy; Secretory Pathway; TA Protein.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures









References
-
- Beilharz T, Egan B, Silver PA, Hofmann K, Lithgow T. Bipartite signals mediate subcellular targeting of tail-anchored membrane proteins in Saccharomyces cerevisiae. J Biol Chem. 2003;278:8219–8223. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

