A compendium of genetic regulatory effects across pig tissues
- PMID: 38177344
- PMCID: PMC10786720
- DOI: 10.1038/s41588-023-01585-7
A compendium of genetic regulatory effects across pig tissues
Abstract
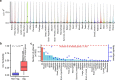
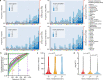
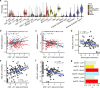
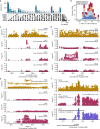
The Farm Animal Genotype-Tissue Expression (FarmGTEx) project has been established to develop a public resource of genetic regulatory variants in livestock, which is essential for linking genetic polymorphisms to variation in phenotypes, helping fundamental biological discovery and exploitation in animal breeding and human biomedicine. Here we show results from the pilot phase of PigGTEx by processing 5,457 RNA-sequencing and 1,602 whole-genome sequencing samples passing quality control from pigs. We build a pig genotype imputation panel and associate millions of genetic variants with five types of transcriptomic phenotypes in 34 tissues. We evaluate tissue specificity of regulatory effects and elucidate molecular mechanisms of their action using multi-omics data. Leveraging this resource, we decipher regulatory mechanisms underlying 207 pig complex phenotypes and demonstrate the similarity of pigs to humans in gene expression and the genetic regulation behind complex phenotypes, supporting the importance of pigs as a human biomedical model.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures












References
MeSH terms
Grants and funding
- MR/R025851/1/RCUK | Medical Research Council (MRC)
- MR/P015514/1/RCUK | Medical Research Council (MRC)
- BBS/E/D/10002070/RCUK | Biotechnology and Biological Sciences Research Council (BBSRC)
- BBS/E/D/30002275/RCUK | Biotechnology and Biological Sciences Research Council (BBSRC)
- 2019-67015-29321/United States Department of Agriculture | Agricultural Research Service (USDA Agricultural Research Service)

