Multi-ancestry genome-wide association study of major depression aids locus discovery, fine mapping, gene prioritization and causal inference
- PMID: 38177345
- PMCID: PMC10864182
- DOI: 10.1038/s41588-023-01596-4
Multi-ancestry genome-wide association study of major depression aids locus discovery, fine mapping, gene prioritization and causal inference
Abstract
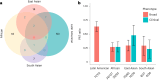
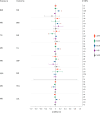
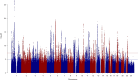
Most genome-wide association studies (GWAS) of major depression (MD) have been conducted in samples of European ancestry. Here we report a multi-ancestry GWAS of MD, adding data from 21 cohorts with 88,316 MD cases and 902,757 controls to previously reported data. This analysis used a range of measures to define MD and included samples of African (36% of effective sample size), East Asian (26%) and South Asian (6%) ancestry and Hispanic/Latin American participants (32%). The multi-ancestry GWAS identified 53 significantly associated novel loci. For loci from GWAS in European ancestry samples, fewer than expected were transferable to other ancestry groups. Fine mapping benefited from additional sample diversity. A transcriptome-wide association study identified 205 significantly associated novel genes. These findings suggest that, for MD, increasing ancestral and global diversity in genetic studies may be particularly important to ensure discovery of core genes and inform about transferability of findings.
© 2024. The Author(s).
Conflict of interest statement
O.G. is now a full-time employee at Union Chimique Belge. A.I.C. is currently an employee of Regeneron Pharmaceuticals and may own stock or stock options. C.T. reported being an employee of and receiving stock options from 23andMe during the conduct of the study. Y.J. reported being an employee of 23andMe outside the submitted work. All other authors declare no competing interests.
Figures






References
-
- GBD 2017 DALYs and HALE Collaborators. Global, regional, and national disability-adjusted life-years (DALYs) for 359 diseases and injuries and healthy life expectancy (HALE) for 195 countries and territories, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet 392, 1859–1922 (2018). - PMC - PubMed
MeSH terms
Grants and funding
- R01 MH100549/MH/NIMH NIH HHS/United States
- N01 HC065233/HL/NHLBI NIH HHS/United States
- S10 RR025141/RR/NCRR NIH HHS/United States
- MC_UU_12026/2/MRC_/Medical Research Council/United Kingdom
- BB/R01356X/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- I01 CX001849/CX/CSRD VA/United States
- R01 MH124873/MH/NIMH NIH HHS/United States
- R01 HD074711/HD/NICHD NIH HHS/United States
- R01 MH125938/MH/NIMH NIH HHS/United States
- N01 HC065237/HL/NHLBI NIH HHS/United States
- MC_U137686851/MRC_/Medical Research Council/United Kingdom
- 210561/Z/18/Z/WT_/Wellcome Trust/United Kingdom
- MR/W014386/1/MRC_/Medical Research Council/United Kingdom
- R01 MD011728/MD/NIMHD NIH HHS/United States
- HHSN268201300005C/HL/NHLBI NIH HHS/United States
- UL1 TR000445/TR/NCATS NIH HHS/United States
- R01 DA009897/DA/NIDA NIH HHS/United States
- 212946/Z/18/Z/WT_/Wellcome Trust/United Kingdom
- R21 DC018098/DC/NIDCD NIH HHS/United States
- UL1 TR001863/TR/NCATS NIH HHS/United States
- N01 HC065236/HL/NHLBI NIH HHS/United States
- N01 HC065235/HL/NHLBI NIH HHS/United States
- U19 HL065962/HL/NHLBI NIH HHS/United States
- R01 MH101459/MH/NIMH NIH HHS/United States
- N01 HC065234/HL/NHLBI NIH HHS/United States
- MC_PC_14135/MRC_/Medical Research Council/United Kingdom
- T32 MH014276/MH/NIMH NIH HHS/United States
- R21 MH126358/MH/NIMH NIH HHS/United States
- RC2 GM092618/GM/NIGMS NIH HHS/United States
- MC_UU_00017/1/MRC_/Medical Research Council/United Kingdom
- P50 GM115305/GM/NIGMS NIH HHS/United States
- P50 HD103537/HD/NICHD NIH HHS/United States
- U01 HG006378/HG/NHGRI NIH HHS/United States
- R33 DA047527/DA/NIDA NIH HHS/United States
- UL1 RR024975/RR/NCRR NIH HHS/United States
- R01 NS032830/NS/NINDS NIH HHS/United States
- U01 HG004798/HG/NHGRI NIH HHS/United States
- UL1 TR002243/TR/NCATS NIH HHS/United States
- R01 MH113930/MH/NIMH NIH HHS/United States

