Disease-associated polyalanine expansion mutations impair UBA6-dependent ubiquitination
- PMID: 38177505
- PMCID: PMC10897158
- DOI: 10.1038/s44318-023-00018-9
Disease-associated polyalanine expansion mutations impair UBA6-dependent ubiquitination
Abstract
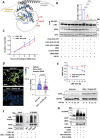
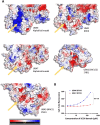
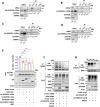
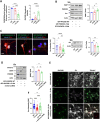
Expansion mutations in polyalanine stretches are associated with a growing number of diseases sharing a high degree of genotypic and phenotypic commonality. These similarities prompted us to query the normal function of physiological polyalanine stretches and to investigate whether a common molecular mechanism is involved in these diseases. Here, we show that UBA6, an E1 ubiquitin-activating enzyme, recognizes a polyalanine stretch within its cognate E2 ubiquitin-conjugating enzyme USE1. Aberrations in this polyalanine stretch reduce ubiquitin transfer to USE1 and, subsequently, polyubiquitination and degradation of its target, the ubiquitin ligase E6AP. Furthermore, we identify competition for the UBA6-USE1 interaction by various proteins with polyalanine expansion mutations in the disease state. The deleterious interactions of expanded polyalanine tract proteins with UBA6 in mouse primary neurons alter the levels and ubiquitination-dependent degradation of E6AP, which in turn affects the levels of the synaptic protein Arc. These effects are also observed in induced pluripotent stem cell-derived autonomic neurons from patients with polyalanine expansion mutations, where UBA6 overexpression increases neuronal resilience to cell death. Our results suggest a shared mechanism for such mutations that may contribute to the congenital malformations seen in polyalanine tract diseases.
Keywords: Autonomic Nervous System; Congenital Central Hypoventilation Syndrome; Trinucleotide Repeats; Ubiquitin Transfer System; Ubiquitin-Activating Enzyme.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Amiel J, Laudier B, Attie-Bitach T, Trang H, de Pontual L, Gener B, Trochet D, Etchevers H, Ray P, Simonneau M, et al. Polyalanine expansion and frameshift mutations of the paired-like homeobox gene PHOX2B in congenital central hypoventilation syndrome. Nat Genet. 2003;33:459–461. doi: 10.1038/ng1130. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

