Shaping immune landscape of colorectal cancer by cholesterol metabolites
- PMID: 38177537
- PMCID: PMC10897227
- DOI: 10.1038/s44321-023-00015-9
Shaping immune landscape of colorectal cancer by cholesterol metabolites
Abstract
Cancer immunotherapies have achieved unprecedented success in clinic, but they remain largely ineffective in some major types of cancer, such as colorectal cancer with microsatellite stability (MSS CRC). It is therefore important to study tumor microenvironment of resistant cancers for developing new intervention strategies. In this study, we identify a metabolic cue that determines the unique immune landscape of MSS CRC. Through secretion of distal cholesterol precursors, which directly activate RORγt, MSS CRC cells can polarize T cells toward Th17 cells that have well-characterized pro-tumor functions in colorectal cancer. Analysis of large human cancer cohorts revealed an asynchronous pattern of the cholesterol biosynthesis in MSS CRC, which is responsible for the abnormal accumulation of distal cholesterol precursors. Inhibiting the cholesterol biosynthesis enzyme Cyp51, by pharmacological or genetic interventions, reduced the levels of intratumoral distal cholesterol precursors and suppressed tumor progression through a Th17-modulation mechanism in preclinical MSS CRC models. Our study therefore reveals a novel mechanism of cancer-immune interaction and an intervention strategy for the difficult-to-treat MSS CRC.
Keywords: Asynchronous Cholesterol Biosynthesis; Colorectal Cancer with Microsatellite Stability; Cyp51 Targeted Therapy; Distal Cholesterol Precursors; Th17.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

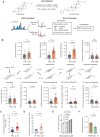

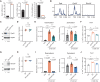

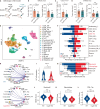

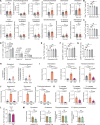

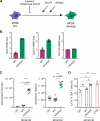
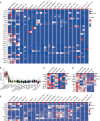
References
-
- Akbay EA, Koyama S, Liu Y, Dries R, Bufe LE, Silkes M, Alam MM, Magee DM, Jones R, Jinushi M, et al. Interleukin-17A promotes lung tumor progression through neutrophil attraction to tumor sites and mediating resistance to PD-1 blockade. J Thorac Oncol. 2017;12:1268–1279. doi: 10.1016/j.jtho.2017.04.017. - DOI - PMC - PubMed
-
- Benjamini Y, Hochberg Y. Controlling the false discovery rate - a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300.
MeSH terms
Grants and funding
- YSBR-014/CAS | Chinese Academy of Sciences Key Project (CAS Key Project)
- 31861133009/MOST | NSFC | Key Programme
- 92057201/MOST | NSFC | Key Programme
- 92157301/MOST | NSFC | Key Programme
- 92169212/MOST | NSFC | Key Programme
- 22022411/MOST | NSFC | Key Programme
- 92057114/MOST | NSFC | Key Programme
- 31971356/MOST | NSFC | Key Programme
- 91857000/MOST | NSFC | Key Programme
- 21JC1405900/STSMC Fundamental Research Program
- 2020000001/CAS Excellent Research Associate Program
- 2020M671260/China Postdoctoral Science Foundation (China Postdoctoral Foundation Project)
- 2022YFC2505004/MOST | National Key Research and Development Program of China (NKPs)
- 2018YFA0800902/MOST | National Key Research and Development Program of China (NKPs)
- 2019SHZDZX02/Shanghai Municipal Science and Technology Major Project
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

