TTLL12 is required for primary ciliary axoneme formation in polarized epithelial cells
- PMID: 38177908
- PMCID: PMC10883266
- DOI: 10.1038/s44319-023-00005-5
TTLL12 is required for primary ciliary axoneme formation in polarized epithelial cells
Abstract
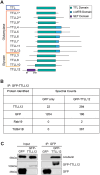
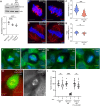
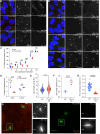
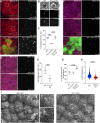
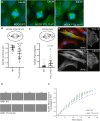
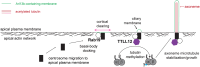
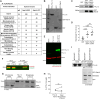
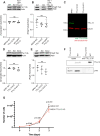
The primary cilium is a critical sensory organelle that is built of axonemal microtubules ensheathed by a ciliary membrane. In polarized epithelial cells, primary cilia reside on the apical surface and must extend these microtubules directly into the extracellular space and remain a stable structure. However, the factors regulating cross-talk between ciliation and cell polarization, as well as axonemal microtubule growth and stabilization in polarized epithelia, are not fully understood. In this study, we find TTLL12, a previously uncharacterized member of the Tubulin Tyrosine Ligase-Like (TTLL) family, localizes to the base of primary cilia and is required for cilia formation in polarized renal epithelial cells. We also show that TTLL12 directly binds to the α/β-tubulin heterodimer in vitro and regulates microtubule dynamics, stability, and post-translational modifications (PTMs). While all other TTLLs catalyze the addition of glutamate or glycine to microtubule C-terminal tails, TTLL12 uniquely affects tubulin PTMs by promoting both microtubule lysine acetylation and arginine methylation. Together, this work identifies a novel microtubule regulator and provides insight into the requirements for apical extracellular axoneme formation.
Keywords: Cilia; Epithelial Cells; Methylation; TTLL12; Tubulin.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Update of
-
TTLL12 is required for primary ciliary axoneme formation in polarized epithelial cells.bioRxiv [Preprint]. 2023 Jul 25:2023.07.25.550533. doi: 10.1101/2023.07.25.550533. bioRxiv. 2023. Update in: EMBO Rep. 2024 Jan;25(1):198-227. doi: 10.1038/s44319-023-00005-5. PMID: 37546873 Free PMC article. Updated. Preprint.
References
-
- Brants J, Semenchenko K, Wasylyk C, Robert A, Carles A, Zambrano A, Pradeau-Aubreton K, Birck C, Schalken JA, Poch O, et al. Tubulin tyrosine ligase like 12, a TTLL family member with SET- and TTL-like domains and roles in histone and tubulin modifications and mitosis. PLoS ONE. 2012;7:e51258. doi: 10.1371/journal.pone.0051258. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

