UBXN1 maintains ER proteostasis and represses UPR activation by modulating translation
- PMID: 38177917
- PMCID: PMC10897191
- DOI: 10.1038/s44319-023-00027-z
UBXN1 maintains ER proteostasis and represses UPR activation by modulating translation
Abstract
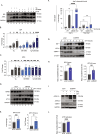
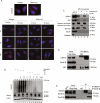
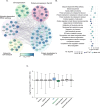
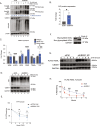
ER protein homeostasis (proteostasis) is essential for proper folding and maturation of proteins in the secretory pathway. Loss of ER proteostasis can lead to the accumulation of misfolded or aberrant proteins in the ER and triggers the unfolded protein response (UPR). In this study, we find that the p97 adaptor UBXN1 is an important negative regulator of the UPR. Loss of UBXN1 sensitizes cells to ER stress and activates the UPR. This leads to widespread upregulation of the ER stress transcriptional program. Using comparative, quantitative proteomics we show that deletion of UBXN1 results in a significant enrichment of proteins involved in ER-quality control processes including those involved in protein folding and import. Notably, we find that loss of UBXN1 does not perturb p97-dependent ER-associated degradation (ERAD). Our studies indicate that loss of UBXN1 increases translation in both resting and ER-stressed cells. Surprisingly, this process is independent of p97 function. Taken together, our studies have identified a new role for UBXN1 in repressing translation and maintaining ER proteostasis in a p97 independent manner.
Keywords: ER Stress; Translation; Ubiquitin; Unfolded Protein Response; p97.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures









References
-
- Atkin JD, Farg MA, Walker AK, Mclean C, Tomas D, Horne MK. Endoplasmic reticulum stress and induction of the unfolded protein response in human sporadic amyotrophic lateral sclerosis. Neurobiol Dis. 2008;30:400–407. - PubMed
-
- Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol. 2006;24:1285–1292. - PubMed
-
- Bellucci A, Navarria L, Zaltieri M, Falarti E, Bodei S, Sigala S, Battistin L, Spillantini M, Missale C, Spano P. Induction of the unfolded protein response by alpha-synuclein in experimental models of Parkinson’s disease. J Neurochem. 2011;116:588–605. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

