BRCC36 Deubiquitinates HMGCR to Regulate the Interplay Between Ferroptosis and Pyroptosis
- PMID: 38178583
- PMCID: PMC10953584
- DOI: 10.1002/advs.202304263
BRCC36 Deubiquitinates HMGCR to Regulate the Interplay Between Ferroptosis and Pyroptosis
Abstract
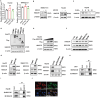
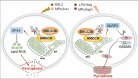
Various forms of programmed cell death (PCD) exhibit distinct characteristics depending on their specific molecular mechanisms, and there are interactions among these different forms. Ferroptosis, which is related to autophagy and apoptosis, has an unknown potential interaction with pyroptosis. This study revealed a mutually antagonistic relationship between ferroptosis and pyroptosis, with 3-hydroxy-3-methylglutaryl-coenzyme A reductase (HMGCR) playing a key role in their interaction. It is found that HMGCR predominantly localized to mitochondria during ferroptosis but shifted to the endoplasmic reticulum following treatment with a pyroptosis inducer. Furthermore, this study demonstrated that BRCC36 (BRCA1/BRCA2-containing complex subunit 36) deubiquitinated HMGCR in a manner dependent on deubiquitinating enzyme (DUB) activity, and inhibited ferroptosis and promoted pyroptosis. Moreover, as an oncogene in hepatocellular carcinoma (HCC), BRCC36 promoted cancer cell proliferation, migration, invasion, and tumor growth. Thiolutin, an inhibitor of BRCC36, effectively suppressed the interaction between BRCC36 and HMGCR, leading to the inhibition of HCC growth. Therefore, targeting BRCC36 can offer a novel and promising therapeutic strategy for HCC treatment. In conclusion, these findings provide new theoretical evidence for further characterizing tumor heterogeneity and offer new molecular targets for the diagnosis and treatment of HCC.
Keywords: BRCC36; HMGCR; endoplasmic reticulum; ferroptosis; mitochondria; pyroptosis.
© 2023 The Authors. Advanced Science published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
- 8237112964/National Natural Science Foundation of China
- 82272982/National Natural Science Foundation of China
- 82073097/National Natural Science Foundation of China
- 81874139/National Natural Science Foundation of China
- 82072594/National Natural Science Foundation of China
- 81872285/National Natural Science Foundation of China
- 81772927/National Natural Science Foundation of China
- Natural Science Foundation of Hunan Province
- 2021SK2013/Hunan Provincial Key Area R&D Programs
- 2023QYJC030/Central South University Research Program of Advanced Interdisciplinary Studies
- 2022RC3072/Science and Technology Innovative Research Team in Higher Educational Institutions of Hunan Province
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
