Integrated single-cell multiomics uncovers foundational regulatory mechanisms of lens development and pathology
- PMID: 38180241
- PMCID: PMC10906490
- DOI: 10.1242/dev.202249
Integrated single-cell multiomics uncovers foundational regulatory mechanisms of lens development and pathology
Abstract
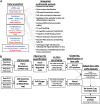
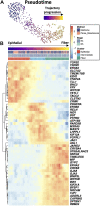
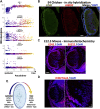
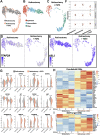
Ocular lens development entails epithelial to fiber cell differentiation, defects in which cause congenital cataracts. We report the first single-cell multiomic atlas of lens development, leveraging snRNA-seq, snATAC-seq and CUT&RUN-seq to discover previously unreported mechanisms of cell fate determination and cataract-linked regulatory networks. A comprehensive profile of cis- and trans-regulatory interactions, including for the cataract-linked transcription factor MAF, is established across a temporal trajectory of fiber cell differentiation. Furthermore, we identify an epigenetic paradigm of cellular differentiation, defined by progressive loss of the H3K27 methylation writer Polycomb repressive complex 2 (PRC2). PRC2 localizes to heterochromatin domains across master-regulator transcription factor gene bodies, suggesting it safeguards epithelial cell fate. Moreover, we demonstrate that FGF hyper-stimulation in vivo leads to MAF network activation and the emergence of novel lens cell states. Collectively, these data depict a comprehensive portrait of lens fiber cell differentiation, while defining regulatory effectors of cell identity and cataract formation.
Keywords: FGF; Lens; MAF; Multiomics; PRC2; Single-cell.
© 2024. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures





Update of
-
Integrated single-cell multiomics uncovers foundational regulatory mechanisms of lens development and pathology.bioRxiv [Preprint]. 2023 Jul 11:2023.07.10.548451. doi: 10.1101/2023.07.10.548451. bioRxiv. 2023. Update in: Development. 2024 Jan 1;151(1):dev202249. doi: 10.1242/dev.202249. PMID: 37502967 Free PMC article. Updated. Preprint.
References
-
- Agrawal, S. A., Anand, D., Siddam, A. D., Kakrana, A., Dash, S., Scheiblin, D. A., Dang, C. A., Terrell, A. M., Waters, S. M., Singh, A.et al. (2015). Compound mouse mutants of bZIP transcription factors Mafg and Mafk reveal a regulatory network of non-crystallin genes associated with cataract. Hum. Genet. 134, 717-735. 10.1007/s00439-015-1554-5 - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

