USP8 prevents aberrant NF-κB and Nrf2 activation by counteracting ubiquitin signals from endosomes
- PMID: 38180476
- PMCID: PMC10783432
- DOI: 10.1083/jcb.202306013
USP8 prevents aberrant NF-κB and Nrf2 activation by counteracting ubiquitin signals from endosomes
Abstract
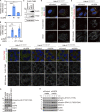
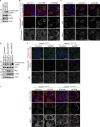
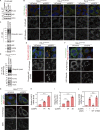
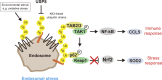
K63-linked ubiquitin chains attached to plasma membrane proteins serve as tags for endocytosis and endosome-to-lysosome sorting. USP8 is an essential deubiquitinase for the maintenance of endosomal functions. Prolonged depletion of USP8 leads to cell death, but the major effects on cellular signaling pathways are poorly understood. Here, we show that USP8 depletion causes aberrant accumulation of K63-linked ubiquitin chains on endosomes and induces immune and stress responses. Upon USP8 depletion, two different decoders for K63-linked ubiquitin chains, TAB2/3 and p62, were recruited to endosomes and activated the TAK1-NF-κB and Keap1-Nrf2 pathways, respectively. Oxidative stress, an environmental stimulus that potentially suppresses USP8 activity, induced accumulation of K63-linked ubiquitin chains on endosomes, recruitment of TAB2, and expression of the inflammatory cytokine. The results demonstrate that USP8 is a gatekeeper of misdirected ubiquitin signals and inhibits immune and stress response pathways by removing K63-linked ubiquitin chains from endosomes.
© 2024 Endo et al.
Conflict of interest statement
Disclosures: The authors declare no competing interests exist.
Figures
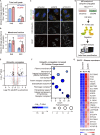
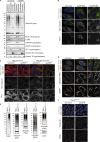

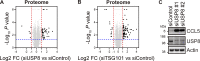
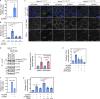


References
-
- Akimov, V., Barrio-Hernandez I., Hansen S.V.F., Hallenborg P., Pedersen A.K., Bekker-Jensen D.B., Puglia M., Christensen S.D.K., Vanselow J.T., Nielsen M.M., et al. . 2018. UbiSite approach for comprehensive mapping of lysine and N-terminal ubiquitination sites. Nat. Struct. Mol. Biol. 25:631–640. 10.1038/s41594-018-0084-y - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

