METTL3 drives telomere targeting of TERRA lncRNA through m6A-dependent R-loop formation: a therapeutic target for ALT-positive neuroblastoma
- PMID: 38180812
- PMCID: PMC10954483
- DOI: 10.1093/nar/gkad1242
METTL3 drives telomere targeting of TERRA lncRNA through m6A-dependent R-loop formation: a therapeutic target for ALT-positive neuroblastoma
Abstract
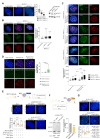
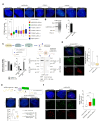
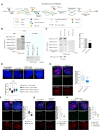
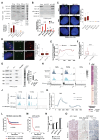
Telomerase-negative tumors maintain telomere length by alternative lengthening of telomeres (ALT), but the underlying mechanism behind ALT remains poorly understood. A proportion of aggressive neuroblastoma (NB), particularly relapsed tumors, are positive for ALT (ALT+), suggesting that a better dissection of the ALT mechanism could lead to novel therapeutic opportunities. TERRA, a long non-coding RNA (lncRNA) derived from telomere ends, localizes to telomeres in a R-loop-dependent manner and plays a crucial role in telomere maintenance. Here we present evidence that RNA modification at the N6 position of internal adenosine (m6A) in TERRA by the methyltransferase METTL3 is essential for telomere maintenance in ALT+ cells, and the loss of TERRA m6A/METTL3 results in telomere damage. We observed that m6A modification is abundant in R-loop enriched TERRA, and the m6A-mediated recruitment of hnRNPA2B1 to TERRA is critical for R-loop formation. Our findings suggest that m6A drives telomere targeting of TERRA via R-loops, and this m6A-mediated R-loop formation could be a widespread mechanism employed by other chromatin-interacting lncRNAs. Furthermore, treatment of ALT+ NB cells with a METTL3 inhibitor resulted in compromised telomere targeting of TERRA and accumulation of DNA damage at telomeres, indicating that METTL3 inhibition may represent a therapeutic approach for ALT+ NB.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



Similar articles
-
METTL3-mediated m6A modification stabilizes TERRA and maintains telomere stability.Nucleic Acids Res. 2022 Nov 11;50(20):11619-11634. doi: 10.1093/nar/gkac1027. Nucleic Acids Res. 2022. PMID: 36399511 Free PMC article.
-
TERRA-LSD1 phase separation promotes R-loop formation for telomere maintenance in ALT cancer cells.Nat Commun. 2024 Mar 9;15(1):2165. doi: 10.1038/s41467-024-46509-z. Nat Commun. 2024. PMID: 38461301 Free PMC article.
-
TERRA and RAD51AP1 promote alternative lengthening of telomeres through an R- to D-loop switch.Mol Cell. 2022 Nov 3;82(21):3985-4000.e4. doi: 10.1016/j.molcel.2022.09.026. Epub 2022 Oct 19. Mol Cell. 2022. PMID: 36265486 Free PMC article.
-
The makings of TERRA R-loops at chromosome ends.Cell Cycle. 2021 Sep;20(18):1745-1759. doi: 10.1080/15384101.2021.1962638. Epub 2021 Aug 25. Cell Cycle. 2021. PMID: 34432566 Free PMC article. Review.
-
TERRA and the alternative lengthening of telomeres: a dangerous affair.FEBS Lett. 2025 Jan;599(2):157-165. doi: 10.1002/1873-3468.14844. Epub 2024 Mar 6. FEBS Lett. 2025. PMID: 38445359 Free PMC article. Review.
Cited by
-
Telomere Maintenance Mechanisms in a Cohort of High-Risk Neuroblastoma Tumors and Its Relation to Genomic Variants in the TERT and ATRX Genes.Cancers (Basel). 2023 Dec 7;15(24):5732. doi: 10.3390/cancers15245732. Cancers (Basel). 2023. PMID: 38136279 Free PMC article.
-
The role of N(6)-methyladenosine (m6a) modification in cancer: recent advances and future directions.EXCLI J. 2025 Jan 15;24:113-150. doi: 10.17179/excli2024-7935. eCollection 2025. EXCLI J. 2025. PMID: 39967906 Free PMC article. Review.
-
Methyltransferase-like 3 is a target for the diagnose and therapy of clear cell renal carcinoma.Front Pharmacol. 2025 Apr 17;16:1534655. doi: 10.3389/fphar.2025.1534655. eCollection 2025. Front Pharmacol. 2025. PMID: 40313614 Free PMC article. Review.
-
TERRA transcripts localize at long telomeres to regulate telomerase access to chromosome ends.Sci Adv. 2024 Jun 14;10(24):eadk4387. doi: 10.1126/sciadv.adk4387. Epub 2024 Jun 12. Sci Adv. 2024. PMID: 38865460 Free PMC article.
-
Dual roles of N6-methyladenosine in R-loop regulation of gene transcription and genome stability.Sci China Life Sci. 2025 Jul 9. doi: 10.1007/s11427-024-2947-6. Online ahead of print. Sci China Life Sci. 2025. PMID: 40643803
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

