Rapid human genomic DNA cloning into mouse artificial chromosome via direct chromosome transfer from human iPSC and CRISPR/Cas9-mediated translocation
- PMID: 38180813
- PMCID: PMC10853801
- DOI: 10.1093/nar/gkad1218
Rapid human genomic DNA cloning into mouse artificial chromosome via direct chromosome transfer from human iPSC and CRISPR/Cas9-mediated translocation
Abstract
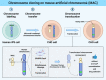
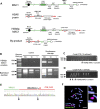
A 'genomically' humanized animal stably maintains and functionally expresses the genes on human chromosome fragment (hCF; <24 Mb) loaded onto mouse artificial chromosome (MAC); however, cloning of hCF onto the MAC (hCF-MAC) requires a complex process that involves multiple steps of chromosome engineering through various cells via chromosome transfer and Cre-loxP chromosome translocation. Here, we aimed to develop a strategy to rapidly construct the hCF-MAC by employing three alternative techniques: (i) application of human induced pluripotent stem cells (hiPSCs) as chromosome donors for microcell-mediated chromosome transfer (MMCT), (ii) combination of paclitaxel (PTX) and reversine (Rev) as micronucleation inducers and (iii) CRISPR/Cas9 genome editing for site-specific translocations. We achieved a direct transfer of human chromosome 6 or 21 as a model from hiPSCs as alternative human chromosome donors into CHO cells containing MAC. MMCT was performed with less toxicity through induction of micronucleation by PTX and Rev. Furthermore, chromosome translocation was induced by simultaneous cleavage between human chromosome and MAC by using CRISPR/Cas9, resulting in the generation of hCF-MAC containing CHO clones without Cre-loxP recombination and drug selection. Our strategy facilitates rapid chromosome cloning and also contributes to the functional genomic analyses of human chromosomes.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




References
-
- Kuroiwa Y., Yoshida H., Ohshima T., Shinohara T., Ohguma A., Kazuki Y., Oshimura M., Ishida I., Tomizuka K.. The use of chromosome-based vectors for animal transgenesis. Gene Ther. 2002; 9:708–712. - PubMed
-
- Tomizuka K., Yoshida H., Uejima H., Kugoh H., Sato K., Ohguma A., Hayasaka M., Hanaoka K., Oshimura M., Ishida I.. Functional expression and germline transmission of a human chromosome fragment in chimaeric mice. Nat. Genet. 1997; 16:133–143. - PubMed
-
- Moriwaki T., Abe S., Oshimura M., Kazuki Y.. Transchromosomic technology for genomically humanized animals. Exp. Cell. Res. 2020; 390:111914. - PubMed
-
- Ma X., Shah Y., Cheung C., Guo G.L., Feigenbaum L., Krausz K.W., Idle J.R., Gonzalez F.J.. The pregnane X receptor gene-humanized mouse: a model for investigating drug-drug interactions mediated by cytochromes P450 3A. Drug Metab. Dispos. 2007; 35:194–200. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

