Impact of systemic antimicrobial therapy on the faecal microbiome in symptomatic dairy cows
- PMID: 38180967
- PMCID: PMC10769045
- DOI: 10.1371/journal.pone.0296290
Impact of systemic antimicrobial therapy on the faecal microbiome in symptomatic dairy cows
Abstract
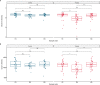
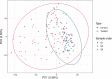
Antimicrobial resistance is a global threat to human and animal health, with the misuse and overuse of antimicrobials suggested as the main drivers of resistance. Antimicrobial therapy can alter the bacterial community composition and the faecal resistome in cattle. Little is known about the impact of systemic antimicrobial therapy on the faecal microbiome in dairy cows in the presence of disease. Therefore, this study aimed to assess the impact of systemic antimicrobial therapy on the faecal microbiome in dairy cows in the pastoral farm environment, by analysing faecal samples from cattle impacted by several different clinically-defined conditions and corresponding antimicrobial treatments. Analysis at the individual animal level showed a decrease in bacterial diversity and richness during antimicrobial treatment but, in many cases, the microbiome diversity recovered post-treatment when the cow re-entered the milking herd. Perturbations in the microbiome composition and the ability of the microbiome to recover were specific at the individual animal level, highlighting that the animal is the main driver of variation. Other factors such as disease severity, the type and duration of antimicrobial treatment and changes in environmental factors may also impact the bovine faecal microbiome. AmpC-producing Escherichia coli were isolated from faeces collected during and post-treatment with ceftiofur from one cow while no third-generation cephalosporin resistant E. coli were isolated from the untreated cow samples. This isolation of genetically similar plasmid-mediated AmpC-producing E. coli has implications for the development and dissemination of antibiotic resistant bacteria and supports the reduction in the use of critically important antimicrobials.
Copyright: © 2024 Collis et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

