Building a vertically integrated genomic learning health system: The biobank at the Colorado Center for Personalized Medicine
- PMID: 38181729
- PMCID: PMC10806731
- DOI: 10.1016/j.ajhg.2023.12.001
Building a vertically integrated genomic learning health system: The biobank at the Colorado Center for Personalized Medicine
Abstract
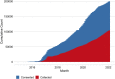
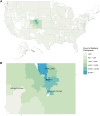
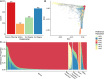
Precision medicine initiatives across the globe have led to a revolution of repositories linking large-scale genomic data with electronic health records, enabling genomic analyses across the entire phenome. Many of these initiatives focus solely on research insights, leading to limited direct benefit to patients. We describe the biobank at the Colorado Center for Personalized Medicine (CCPM Biobank) that was jointly developed by the University of Colorado Anschutz Medical Campus and UCHealth to serve as a unique, dual-purpose research and clinical resource accelerating personalized medicine. This living resource currently has more than 200,000 participants with ongoing recruitment. We highlight the clinical, laboratory, regulatory, and HIPAA-compliant informatics infrastructure along with our stakeholder engagement, consent, recontact, and participant engagement strategies. We characterize aspects of genetic and geographic diversity unique to the Rocky Mountain region, the primary catchment area for CCPM Biobank participants. We leverage linked health and demographic information of the CCPM Biobank participant population to demonstrate the utility of the CCPM Biobank to replicate complex trait associations in the first 33,674 genotyped individuals across multiple disease domains. Finally, we describe our current efforts toward return of clinical genetic test results, including high-impact pathogenic variants and pharmacogenetic information, and our broader goals as the CCPM Biobank continues to grow. Bringing clinical and research interests together fosters unique clinical and translational questions that can be addressed from the large EHR-linked CCPM Biobank resource within a HIPAA- and CLIA-certified environment.
Keywords: biobanking; electronic health records; learning health system; pharmacogenomics; precision medicine.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests K.C.B. owns stock in Tempus and Galatea Bio and is an employee of Oxford Nanopore Technologies. C.R.G. owns stock in 23andMe, Inc.
Figures


References
-
- Carey D.J., Fetterolf S.N., Davis F.D., Faucett W.A., Kirchner H.L., Mirshahi U., Murray M.F., Smelser D.T., Gerhard G.S., Ledbetter D.H. The Geisinger MyCode community health initiative: an electronic health record-linked biobank for precision medicine research. Genet. Med. 2016;18:906–913. - PMC - PubMed
-
- Dewey F.E., Murray M.F., Overton J.D., Habegger L., Leader J.B., Fetterolf S.N., O'Dushlaine C., Van Hout C.V., Staples J., Gonzaga-Jauregui C., et al. Distribution and clinical impact of functional variants in 50,726 whole-exome sequences from the DiscovEHR study. Science. 2016;354 - PubMed
Publication types
MeSH terms
Grants and funding
- UM1 AI109565/AI/NIAID NIH HHS/United States
- R25 HL146166/HL/NHLBI NIH HHS/United States
- UL1 TR002535/TR/NCATS NIH HHS/United States
- R01 HG011345/HG/NHGRI NIH HHS/United States
- R01 HL151152/HL/NHLBI NIH HHS/United States
- K01 LM013088/LM/NLM NIH HHS/United States
- U19 AI117673/AI/NIAID NIH HHS/United States
- U01 HG011715/HG/NHGRI NIH HHS/United States
- R01 HL104608/HL/NHLBI NIH HHS/United States
- R01 AI132476/AI/NIAID NIH HHS/United States
- UM1 TR004399/TR/NCATS NIH HHS/United States
- R01 HG010067/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources

