Chronic hypoxia stabilizes 3βHSD1 via autophagy suppression
- PMID: 38181788
- PMCID: PMC10851248
- DOI: 10.1016/j.celrep.2023.113575
Chronic hypoxia stabilizes 3βHSD1 via autophagy suppression
Abstract
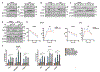
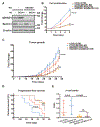
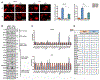
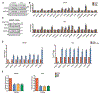
Progression of prostate cancer depends on androgen receptor, which is usually activated by androgens. Therefore, a mainstay treatment is androgen deprivation therapy. Unfortunately, despite initial treatment response, resistance nearly always develops, and disease progresses to castration-resistant prostate cancer (CRPC), which remains driven by non-gonadal androgens synthesized in prostate cancer tissues. 3β-Hydroxysteroid dehydrogenase/Δ5-->4 isomerase 1 (3βHSD1) catalyzes the rate-limiting step in androgen synthesis. However, how 3βHSD1, especially the "adrenal-permissive" 3βHSD1(367T) that permits tumor synthesis of androgen from dehydroepiandrosterone (DHEA), is regulated at the protein level is not well understood. Here, we investigate how hypoxia regulates 3βHSD1(367T) protein levels. Our results show that, in vitro, hypoxia stabilizes 3βHSD1 protein by suppressing autophagy. Autophagy inhibition promotes 3βHSD1-dependent tumor progression. Hypoxia represses transcription of autophagy-related (ATG) genes by decreasing histone acetylation. Inhibiting deacetylase (HDAC) restores ATG gene transcription under hypoxia. Therefore, HDAC inhibition may be a therapeutic target for hypoxic tumor cells.
Keywords: 3βHSD1; CP: Cancer; CP: Molecular biology; androgen synthesis; autophagy; enzyme; germline; hypoxia; metabolism; prostate cancer; protein; steroid.
Copyright © 2023 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests N.S. reports grants from BMS and grants from Astellas outside the submitted work; in addition, N.S. has a patent for HSD3B1 in prostate cancer issued.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

