Boosting with adjuvanted SCB-2019 elicits superior Fcγ-receptor engagement driven by IgG3 to SARS-CoV-2 spike
- PMID: 38182593
- PMCID: PMC10770118
- DOI: 10.1038/s41541-023-00791-y
Boosting with adjuvanted SCB-2019 elicits superior Fcγ-receptor engagement driven by IgG3 to SARS-CoV-2 spike
Abstract
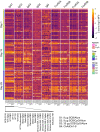
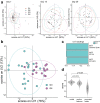
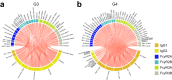
With the continued emergence of variants of concern, the global threat of COVID-19 persists, particularly in low- and middle-income countries with limited vaccine access. Protein-based vaccines, such as SCB-2019, can be produced on a large scale at a low cost while antigen design and adjuvant use can modulate efficacy and safety. While effective humoral immunity against SARS-CoV-2 variants has been shown to depend on both neutralization and Fc-mediated immunity, data on the effectiveness of protein-based vaccines with enhanced Fc-mediated immunity is limited. Here, we assess the humoral profile, including antibody isotypes, subclasses, and Fc receptor binding generated by a boosting with a recombinant trimer-tag protein vaccine SCB-2019. Individuals who were primed with 2 doses of the ChAdOx1 vaccine were equally divided into 4 groups and boosted with following formulations: Group 1: 9 μg SCB-2019 and Alhydrogel; Group 2: 9 μg SCB-2019, CpG 1018, and Alhydrogel; Group 3: 30 μg SCB-2019, CpG 1018, and Alhydrogel; Group 4: ChAdOx1. Group 3 showed enhanced antibody FcγR binding against wild-type and variants compared to Groups 1 and 2, showing a dose-dependent enhancement of immunity conferred by the SCB-2019 vaccine. Moreover, from day 15 after vaccination, Group 3 exhibited higher IgG3 and FcγR binding across variants of concerns, including Omicron and its subvariants, compared to the ChAdOx1-boosted individuals. Overall, this highlights the potential of SCB-2019 as a cost-efficient boosting regimen effective across variants of concerns.
© 2024. The Author(s).
Conflict of interest statement
G.A. is an employee of Moderna Therapeutics and holds equity in Leyden Labs and Seromyx Systems. I.S. is a full-time employee of Clover Biopharmaceuticals. R.C. is a scientific adviser for Clover Biopharmaceuticals. S.A.C.C. is a member of the Oxford Vaccine Group and was involved in the development of the ChadOx-1 vaccine; she is also a scientific adviser for Clover Biopharmaceuticals.
Figures


References
-
- WHO Coronavirus (COVID-19) Dashboard. https://covid19.who.int/?mapFilter=cases (2023).
LinkOut - more resources
Full Text Sources
Miscellaneous

