Ripretinib versus sunitinib in gastrointestinal stromal tumor: ctDNA biomarker analysis of the phase 3 INTRIGUE trial
- PMID: 38182785
- PMCID: PMC10878977
- DOI: 10.1038/s41591-023-02734-5
Ripretinib versus sunitinib in gastrointestinal stromal tumor: ctDNA biomarker analysis of the phase 3 INTRIGUE trial
Abstract
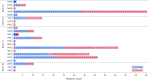
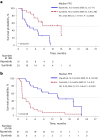
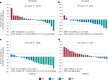
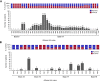
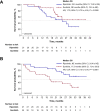
INTRIGUE was an open-label, phase 3 study in adult patients with advanced gastrointestinal stromal tumor who had disease progression on or intolerance to imatinib and who were randomized to once-daily ripretinib 150 mg or sunitinib 50 mg. In the primary analysis, progression-free survival (PFS) with ripretinib was not superior to sunitinib. In clinical and nonclinical studies, ripretinib and sunitinib have demonstrated differential activity based on the exon location of KIT mutations. Therefore, we hypothesized that mutational analysis using circulating tumor DNA (ctDNA) might provide further insight. In this exploratory analysis (N = 362), baseline peripheral whole blood was analyzed by a 74-gene ctDNA next-generation sequencing-based assay. ctDNA was detected in 280/362 (77%) samples with KIT mutations in 213/362 patients (59%). Imatinib-resistant mutations were found in the KIT ATP-binding pocket (exons 13/14) and activation loop (exons 17/18). Mutational subgroup assessment showed 2 mutually exclusive populations with differential treatment effects. Patients with only KIT exon 11 + 13/14 mutations (ripretinib, n = 21; sunitinib, n = 20) had better PFS with sunitinib versus ripretinib (median, 15.0 versus 4.0 months). Patients with only KIT exon 11 + 17/18 mutations (ripretinib, n = 27; sunitinib, n = 25) had better PFS with ripretinib versus sunitinib (median, 14.2 versus 1.5 months). The results of this exploratory analysis suggest ctDNA sequencing may improve the prediction of the efficacy of single-drug therapies and support further evaluation of ripretinib in patients with KIT exon 11 + 17/18 mutations. ClinicalTrials.gov identifier: NCT03673501.
© 2024. The Author(s).
Conflict of interest statement
M.C.H. reports honoraria from Novartis; consulting/advisory roles for Blueprint Medicines, Deciphera, Novartis and Theseus Pharmaceuticals; patents/royalties/other intellectual property licensed to Novartis (institutional (Inst); treatment of GIST); and partial salary from a research grant from the Jonathan David Foundation, a VA Merit Review Grant (I01BX005358) and NCI R21 grant (R21CA263400). R.L.J. reports consulting/advisory roles for Lilly, Immune Design, Merck Serono, Adaptimmune, Daiichi Sankyo, Eisai, Morphotek, TRACON Pharma, Immodulon Therapeutics, Deciphera Pharmaceuticals, PharmaMar, Blueprint Medicines, Clinigen Group, Epizyme, Boehringer Ingelheim, Bayer, Karma Oncology, UpToDate, Athenex, Immunicum, Mundipharma, SpringWorks Therapeutics and SynOX; funding for travel/accommodations/expenses from PharmaMar; and research funding from GlaxoSmithKline (Inst). S.G. reports stock/other ownership interests from Abbott Laboratories; honoraria from CStone Pharmaceuticals; consulting or advisory roles for Blueprint Medicines, Deciphera, Bayer, Lilly, UpToDate, Research to Practice, MORE Health, Daiichi, KayoThera and Immunicum; research funding from Blueprint Medicines (Inst), Deciphera (Inst), Daiichi Sankyo RD Novare (Inst), Merck (Inst), Eisai (Inst), SpringWorks Therapeutics (Inst), TRACON Pharma (Inst), Theseus Pharmaceuticals (Inst) and BioAtla; patents/royalties/other intellectual property from UpToDate; expert testimony for Bayer; and other relationships with Research to Practice and WCG. H.G. reports research funding from Novartis (Inst), Ipsen (Inst), Deciphera (Inst) and Daiichi Sankyo (Inst). P.S. reports honoraria from Blueprint Medicines; consulting/advisory roles for Deciphera, Ellipses Pharma, Blueprint Medicines, Transgene, Exelixis, Boehringer Ingelheim, Studiecentrum voor Kernenergie (SCK CEN), SQZ Biotechnology, Adcendo, PharmaMar, Merck and Medpace; and research funding from CoBioRes NV (Inst), Eisai (Inst), G1 Therapeutics (Inst), PharmaMar (Inst), Genmab (Inst), Merck (Inst), Sartar Therapeutics (Inst) and ONA Therapeutics (Inst). M.v.M. reports consulting/advisory roles for Deciphera and GlaxoSmithKline; research funding from Novartis (Inst), Deciphera (Inst), Gradalis (Inst), Genmab (Inst), ASCO (Inst) and Solaris Health (Inst); patents/royalties/other intellectual property from Cell Line; and a relationship with the National Comprehensive Cancer Network. J.R.Z. reports a leadership role with ICON Group; stock/other ownership interests in Biomarin, Opthea, Amarin Corporation, Concert Pharmaceuticals, Frequency Therapeutics, Gilead Sciences, Madrigal Pharmaceuticals, UniQure, Zogenix, Orphazyme, Moderna Therapeutics, Twist Bioscience and Novavax; honoraria from Specialised Therapeutics, Merck Serono, Targovax, Halozyme, Gilead Sciences and Deciphera; consulting/advisory roles with Merck Serono, Targovax, Merck Sharp & Dohme, Halozyme, Lipotek, Specialised Therapeutics, Center for Emerging & Neglected Diseases (CEND), Deciphera, REVOLUTION MEDICINE, FivePHusion, Genor BioPharma, 1Globe Health Institute and Novotech; research funding from Merck Serono (Inst), Bristol Myers Squibb (Inst), AstraZeneca (Inst), Pfizer (Inst), IQVIA (Inst), Mylan (Inst), Ipsen (Inst), Eisai (Inst), Medtronic (Inst) and MSD Oncology (Inst); and funding for travel/accommodations/expenses from Merck Serono, AstraZeneca, Merck Sharp & Dohme, Deciphera and Sanofi. Y.-K.K. reports consulting/advisory roles with DAEHWA Pharmaceutical, Bristol Myers Squibb, Zymeworks, ALX Oncology, Amgen, Novartis, Macrogenics, Surface Oncology and Blueprint Medicines. A.A.R. reports consulting/advisory roles with Adaptimmune, Bayer, GlaxoSmithKline, Medison and Inhibrx; research funding from Deciphera, Karyopharm Therapeutics, Pfizer, Roche/Genentech, Bristol Myers Squibb, MedImmune, Amgen, GlaxoSmithKline, Blueprint Medicines, Merck, AbbVie, Adaptimmune, Iterion Therapeutics, Neoleukin Therapeutics, Daiichi Sankyo, Symphogen and Rain Therapeutics; and expert testimony for Medison. J.T. reports consulting/advisory roles for Blueprint Medicines, Deciphera, Daiichi Sankyo, Epizyme, Agios, C4 Therapeutics, Bayer, AADI Bioscience, LLC, Foghorn Therapeutics, Boehringer Ingelheim, Cogent Medicine and SERVIER. S.A. reports research funding from AB Science (Inst), TRACON Pharma (Inst), Bayer (Inst), Novartis (Inst), Lilly (Inst), Immune Design (Inst), Karyopharm Therapeutics (Inst), Epizyme (Inst), Blueprint Medicines (Inst), Genmab (Inst), CBA Pharma (Inst), Desmoid Tumor Research Foundation, Merck (Inst), Philogen (Inst), Gradalis (Inst), Deciphera (Inst), Takeda (Inst), Incyte (Inst), SpringWorks Therapeutics (Inst), Adaptimmune (Inst), Advenchen Laboratories (Inst), Bavarian Nordic (Inst), BTG (Inst), PTC Therapeutics (Inst), GlaxoSmithKline (Inst), FORMA Therapeutics (Inst), TRACON Pharma (Inst), Ayala Pharmaceuticals (Inst), Trillium Therapeutics (Inst), Boehringer Ingelheim (Inst), Salarius Pharmaceuticals (Inst), Theseus Pharmaceuticals (Inst), Monopar Therapeutics (Inst), C4 Therapeutics (Inst), InhibRx (Inst), Noxopharm (Inst) and Rain Therapeutics (Inst). A.L.C. reports honoraria from Bayer, PharmaMar and Deciphera. B.L.S. reports a consulting/advisory role for Deciphera (Inst); and research funding from Deciphera (Inst). D.G. reports honoraria from Sun Biopharma, Boehringer Ingelheim and AstraZeneca; consulting/advisory roles for Sun Biopharma, Seattle Genetics, AstraZeneca and Boehringer Ingelheim; and research funding from Amgen (Inst), Pfizer (Inst), Celgene (Inst), Bayer (Inst), Zucero Therapeutics (Inst) and Bristol Myers Squibb (Inst). K.B. reports honoraria from Novartis; and a consulting/advisory role for GlaxoSmithKline. C.S. reports research funding from Roche and Novartis; expert testimony for Pfizer; and funding for travel/accommodations/expenses from Novartis and Pfizer. N.S. reports consulting/advisory roles for Boehringer Ingelheim (Inst), Ellipses Pharma (Inst), Cogent Biosciences (Inst) and Luszana (Inst); and research funding from AstraZeneca/MedImmune (Inst), Bayer (Inst), Bristol Myers Squibb (Inst), Novartis (Inst), GlaxoSmithKline (Inst), Pfizer (Inst), Roche (Inst), Boehringer Ingelheim (Inst), Blueprint Medicines (Inst), Deciphera (Inst), Genentech (Inst), Merck Sharp & Dohme (Inst), Amgen (Inst), Merus (Inst), Incyte (Inst), AbbVie (Inst), Actuate Therapeutics (Inst), Sanofi (Inst), Cytovation (Inst), InteRNA (Inst), Array BioPharma (Inst), Cantargia AB (Inst), Taiho Pharmaceutical (Inst), Takeda (Inst), Ascendis Pharma (Inst), BridgeBio Pharma (Inst), CellCentric (Inst), Crescendo Biologics (Inst), Lilly (Inst), Exelixis (Inst), Janssen (Inst), Merck KGaA (Inst), Molecular Partners (Inst), Numab (Inst), Seattle Genetics (Inst), Cogent Biosciences (Inst), Kinnate Biopharma (Inst), Kling Biotherapeutics (Inst), Navire (Inst), Luszana (Inst), Relay Therapeutics (Inst), Revolution Medicines (Inst) and Dragonfly Therapeutics (Inst). P.R. reports honoraria from MSD, BMS and Pierre Fabre; advisory board roles for MSD, BMS, Pierre Fabre, Merck, Sanofi, Blueprint Medicines and Philogen; invited speaker roles for Merck, Sanofi and Novartis; research funding from Pfizer (Inst) and BMS (Inst); and nonfinancial interests as an invited speaker for the Polish Society of Surgical Oncology and the Polish Oncological Society (President Elect), and as an officer for ASCO. M.D. reports consulting/advisory roles for Adaptimmune, Deciphera and Epizyme; and speakers’ bureau participation for Deciphera. C.S. reports consulting/advisory roles for Deciphera, Blueprint Medicines, Immunicum and Cogent Biosciences; funding for travel/accommodations/expenses from PharmaMar, Pfizer, Bayer and Gilead Sciences; honoraria from Deciphera and PharmaMar; and research funding from Karyopharm Therapeutics (Inst) and IDRX (Inst). N.S. reports consulting/advisory roles for Deciphera, AADi, Blueprint Medicines, Bayer, Epizyme and Boehringer Ingelheim; stock/other ownership interests in Pfizer and Johnson & Johnson; and research funding from GlaxoSmithKline, Karyopharm Therapeutics, Deciphera, Ascentage Pharma, Daiichi Sankyo/Lilly and AstraZeneca/MedImmune (Inst). P.C. reports consulting/advisory roles for Deciphera, NewBay Pharma and Zai Lab; patents/royalties/other intellectual property from ORIC pharmaceuticals (immediate family member); stock/other ownership interests in ORIC pharmaceuticals (immediate family member); and research funding from Deciphera (Inst), Pfizer (Inst) and NewBay Pharma (Inst). W.R. reports employment with Deciphera; and stock and other ownership interests in Deciphera. K.S. reports employment with Deciphera (self) and Stablix (immediate family member); stock and other ownership interests in Deciphera (self) and Stablix (immediate family member); and a consulting/advisory role for Ipsen (immediate family member). H.A. reports employment with Deciphera; and stock and other ownership interests in Deciphera. M.L.S. reports employment with Deciphera; independent board of director position with Pieris Pharmaceuticals; leadership roles with Deciphera and Pieris Pharmaceuticals; stock/other ownership interests in Deciphera and Pieris Pharmaceuticals; and patents/royalties/other intellectual property from Acceleron Pharma. R.R.-S. reports employment with Deciphera; stock/other ownership interests in Deciphera and Immunogen; and patents/royalties/other intellectual property from Immunogen (inventor in three patents with Immunogen, transferred the rights to Immunogen, has not received (and will not receive) any royalties) and Deciphera (inventor in pending patents at Deciphera, transferred the rights to Deciphera, has not received (and will not receive) any royalties). J.-Y.B. reports a leadership role with Innate Pharma; honoraria from Roche, AstraZeneca, PharmaMar, MSD, BMS, Bayer, Ignyta, and Deciphera; consulting/advisory roles with Roche, PharmaMar, Blueprint Medicines, Bayer, Deciphera and Karyopharm Therapeutics; research funding from GlaxoSmithKline (Inst), PharmaMar (Inst), Novartis (Inst), Bayer (Inst), Roche (Inst), BMS (Inst), MSD (Inst), Deciphera (Inst), AstraZeneca (Inst) and OSE Pharma (Inst); and funding for travel/accommodations/expenses from Roche. S.B. reports honoraria from Novartis, Pfizer, Bayer, PharmaMar, GlaxoSmithKline and Deciphera; consulting/advisory roles with Blueprint Medicines, Bayer, Lilly, Deciphera, Nanobiotix, Daiichi Sankyo, Exelixis, Janssen-Cilag, ADC Therapeutics, Mundipharma, GlaxoSmithKline, Adcendo and Boehringer Ingelheim; research funding from Blueprint Medicines, Novartis and Incyte (Inst); and funding for travel/accommodations/expenses from PharmaMar.
Figures


Comment in
-
Selection between sunitinib and ripretinib using mutation analysis for gastrointestinal stromal tumor patients progressing on imatinib.Transl Gastroenterol Hepatol. 2024 Sep 11;9:52. doi: 10.21037/tgh-24-32. eCollection 2024. Transl Gastroenterol Hepatol. 2024. PMID: 39503033 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

