LDLR is an entry receptor for Crimean-Congo hemorrhagic fever virus
- PMID: 38182887
- PMCID: PMC10837205
- DOI: 10.1038/s41422-023-00917-w
LDLR is an entry receptor for Crimean-Congo hemorrhagic fever virus
Abstract
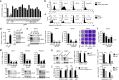
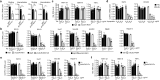
Crimean-Congo hemorrhagic fever virus (CCHFV) is the most widespread tick-born zoonotic bunyavirus that causes severe hemorrhagic fever and death in humans. CCHFV enters the cell via clathrin-mediated endocytosis which is dependent on its surface glycoproteins. However, the cellular receptors that are required for CCHFV entry are unknown. Here we show that the low density lipoprotein receptor (LDLR) is an entry receptor for CCHFV. Genetic knockout of LDLR impairs viral infection in various CCHFV-susceptible human, monkey and mouse cells, which is restored upon reconstitution with ectopically-expressed LDLR. Mutagenesis studies indicate that the ligand binding domain (LBD) of LDLR is necessary for CCHFV infection. LDLR binds directly to CCHFV glycoprotein Gc with high affinity, which supports virus attachment and internalization into host cells. Consistently, a soluble sLDLR-Fc fusion protein or anti-LDLR blocking antibodies impair CCHFV infection into various susceptible cells. Furthermore, genetic knockout of LDLR or administration of an LDLR blocking antibody significantly reduces viral loads, pathological effects and death following CCHFV infection in mice. Our findings suggest that LDLR is an entry receptor for CCHFV and pharmacological targeting of LDLR may provide a strategy to prevent and treat Crimean-Congo hemorrhagic fever.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Crimean-Congo haemorrhagic fever virus uses LDLR to bind and enter host cells.Nat Microbiol. 2024 Jun;9(6):1499-1512. doi: 10.1038/s41564-024-01672-3. Epub 2024 Mar 28. Nat Microbiol. 2024. PMID: 38548922 Free PMC article.
-
Structure and Characterization of Crimean-Congo Hemorrhagic Fever Virus GP38.J Virol. 2020 Mar 31;94(8):e02005-19. doi: 10.1128/JVI.02005-19. Print 2020 Mar 31. J Virol. 2020. PMID: 31996434 Free PMC article.
-
Nucleoside-Modified mRNA Vaccines Protect IFNAR-/- Mice against Crimean-Congo Hemorrhagic Fever Virus Infection.J Virol. 2022 Feb 9;96(3):e0156821. doi: 10.1128/JVI.01568-21. Epub 2021 Nov 24. J Virol. 2022. PMID: 34817199 Free PMC article.
-
Molecular Insights into Crimean-Congo Hemorrhagic Fever Virus.Viruses. 2016 Apr 21;8(4):106. doi: 10.3390/v8040106. Viruses. 2016. PMID: 27110812 Free PMC article. Review.
-
Crimean-Congo hemorrhagic fever: history, epidemiology, pathogenesis, clinical syndrome and genetic diversity.Antiviral Res. 2013 Oct;100(1):159-89. doi: 10.1016/j.antiviral.2013.07.006. Epub 2013 Jul 29. Antiviral Res. 2013. PMID: 23906741 Review.
Cited by
-
Viral hijacking of host DDX60 promotes Crimean-Congo haemorrhagic fever virus replication via G-quadruplex unwinding.PLoS Pathog. 2025 Jun 27;21(6):e1013278. doi: 10.1371/journal.ppat.1013278. eCollection 2025 Jun. PLoS Pathog. 2025. PMID: 40577441 Free PMC article.
-
Engineering and structures of Crimean-Congo hemorrhagic fever virus glycoprotein complexes.Cell. 2025 Jan 23;188(2):303-315.e13. doi: 10.1016/j.cell.2024.11.008. Epub 2024 Dec 18. Cell. 2025. PMID: 39701101
-
Avidity and variable domain spacing strongly influence the therapeutic potency of bispecific antibodies against Crimean-Congo hemorrhagic fever virus.mBio. 2025 May 14;16(5):e0320224. doi: 10.1128/mbio.03202-24. Epub 2025 Apr 16. mBio. 2025. PMID: 40237506 Free PMC article.
-
Recent Advances in Crimean-Congo Hemorrhagic Fever Virus Detection, Treatment, and Vaccination: Overview of Current Status and Challenges.Biol Proced Online. 2024 Jun 26;26(1):20. doi: 10.1186/s12575-024-00244-3. Biol Proced Online. 2024. PMID: 38926669 Free PMC article. Review.
-
Lrp1 facilitates infection of neurons by Jamestown Canyon virus.bioRxiv [Preprint]. 2024 Nov 6:2024.11.06.622176. doi: 10.1101/2024.11.06.622176. bioRxiv. 2024. PMID: 39574651 Free PMC article. Preprint.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

