The potential of epigenetic therapy to target the 3D epigenome in endocrine-resistant breast cancer
- PMID: 38182927
- PMCID: PMC10948365
- DOI: 10.1038/s41594-023-01181-7
The potential of epigenetic therapy to target the 3D epigenome in endocrine-resistant breast cancer
Abstract
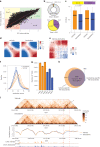
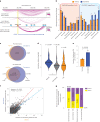
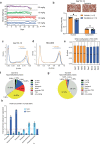
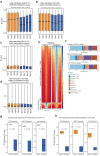
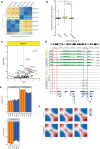
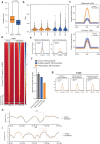
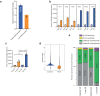
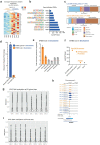
Three-dimensional (3D) epigenome remodeling is an important mechanism of gene deregulation in cancer. However, its potential as a target to counteract therapy resistance remains largely unaddressed. Here, we show that epigenetic therapy with decitabine (5-Aza-mC) suppresses tumor growth in xenograft models of pre-clinical metastatic estrogen receptor positive (ER+) breast tumor. Decitabine-induced genome-wide DNA hypomethylation results in large-scale 3D epigenome deregulation, including de-compaction of higher-order chromatin structure and loss of boundary insulation of topologically associated domains. Significant DNA hypomethylation associates with ectopic activation of ER-enhancers, gain in ER binding, creation of new 3D enhancer-promoter interactions and concordant up-regulation of ER-mediated transcription pathways. Importantly, long-term withdrawal of epigenetic therapy partially restores methylation at ER-enhancer elements, resulting in a loss of ectopic 3D enhancer-promoter interactions and associated gene repression. Our study illustrates the potential of epigenetic therapy to target ER+ endocrine-resistant breast cancer by DNA methylation-dependent rewiring of 3D chromatin interactions, which are associated with the suppression of tumor growth.
© 2024. The Author(s).
Conflict of interest statement
A.S. is an employee of Arima Genomics. The other authors declare no competing interests.
Figures








References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

