Identification of genes regulated by 20-Hydroxyecdysone in Macrobrachium nipponense using comparative transcriptomic analysis
- PMID: 38183039
- PMCID: PMC10768235
- DOI: 10.1186/s12864-023-09927-9
Identification of genes regulated by 20-Hydroxyecdysone in Macrobrachium nipponense using comparative transcriptomic analysis
Abstract
Background: Macrobrachium nipponense is a freshwater prawn of economic importance in China. Its reproductive molt is crucial for seedling rearing and directly impacts the industry's economic efficiency. 20-hydroxyecdysone (20E) controls various physiological behaviors in crustaceans, among which is the initiation of molt. Previous studies have shown that 20E plays a vital role in regulating molt and oviposition in M. nipponense. However, research on the molecular mechanisms underlying the reproductive molt and role of 20E in M. nipponense is still limited.
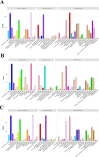
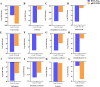
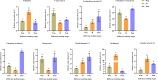
Results: A total of 240.24 Gb of data was obtained from 18 tissue samples by transcriptome sequencing, with > 6 Gb of clean reads per sample. The efficiency of comparison with the reference transcriptome ranged from 87.05 to 92.48%. A total of 2532 differentially expressed genes (DEGs) were identified. Eighty-seven DEGs associated with molt or 20E were screened in the transcriptomes of the different tissues sampled in both the experimental and control groups. The reliability of the RNA sequencing data was confirmed using Quantitative Real-Time PCR. The expression levels of the eight strong candidate genes showed significant variation at the different stages of molt.
Conclusion: This study established the first transcriptome library for the different tissues of M. nipponense in response to 20E and demonstrated the dominant role of 20E in the molting process of this species. The discovery of a large number of 20E-regulated strong candidate DEGs further confirms the extensive regulatory role of 20E and provides a foundation for the deeper understanding of its molecular regulatory mechanisms.
Keywords: 20-hydroxyecdysone; Macrobrachium nipponense; Molting; Transcriptome.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
MnHR4 Functions during Molting of Macrobrachium nipponense by Regulating 20E Synthesis and Mediating 20E Signaling.Int J Mol Sci. 2022 Oct 19;23(20):12528. doi: 10.3390/ijms232012528. Int J Mol Sci. 2022. PMID: 36293382 Free PMC article.
-
RNA interference shows that Spook, the precursor gene of 20-hydroxyecdysone (20E), regulates the molting of Macrobrachium nipponense.J Steroid Biochem Mol Biol. 2021 Oct;213:105976. doi: 10.1016/j.jsbmb.2021.105976. Epub 2021 Aug 18. J Steroid Biochem Mol Biol. 2021. PMID: 34418528
-
Deciphering Molecular Mechanisms Governing the Reproductive Molt of Macrobrachium nipponense: A Transcriptome Analysis of Ovaries across Various Molting Stages.Int J Mol Sci. 2023 Jul 4;24(13):11056. doi: 10.3390/ijms241311056. Int J Mol Sci. 2023. PMID: 37446235 Free PMC article.
-
MnFtz-f1 Is Required for Molting and Ovulation of the Oriental River Prawn Macrobrachium nipponense.Front Endocrinol (Lausanne). 2021 Dec 20;12:798577. doi: 10.3389/fendo.2021.798577. eCollection 2021. Front Endocrinol (Lausanne). 2021. PMID: 34987481 Free PMC article.
-
Combined transcriptome and metabolome analysis reveal key regulatory genes and pathways of feed conversion efficiency of oriental river prawn Macrobrachium nipponense.BMC Genomics. 2023 May 19;24(1):267. doi: 10.1186/s12864-023-09317-1. BMC Genomics. 2023. PMID: 37208591 Free PMC article.
Cited by
-
Genome sequencing of Caridina pseudogracilirostris and its comparative analysis with malacostracan crustaceans.3 Biotech. 2024 Nov;14(11):276. doi: 10.1007/s13205-024-04121-4. Epub 2024 Oct 23. 3 Biotech. 2024. PMID: 39464522
-
Comparative Transcriptome Analysis of Hepatopancreas Reveals Sexual Dimorphic Response to Methyl Farnesoate Injection in Litopenaeus vannamei.Int J Mol Sci. 2024 Jul 26;25(15):8152. doi: 10.3390/ijms25158152. Int J Mol Sci. 2024. PMID: 39125723 Free PMC article.
References
-
- Petryk A, Warren JT, Marqués G, Jarcho MP, Gilbert LI, Kahler J, et al. Shade is the Drosophila P450 enzyme that mediates the hydroxylation of ecdysone to the steroid insect molting hormone 20-hydroxyecdysone. Proc Natl Acad Sci USA. 2003;100(24):13773–13778. doi: 10.1073/pnas.2336088100. - DOI - PMC - PubMed
-
- Swevers L, Iatrou K. Ecdysone Struct Funct. 2009. Ecdysteroids and ecdysteroid signaling pathways during insect oogenesis; pp. 127–64.
-
- Baldini F, Gabrieli P, South A, Valim C, Mancini F, Catteruccia F. The interaction between a sexually transferred steroid hormone and a female protein regulates oogenesis in the Malaria mosquito Anopheles gambiae. PloS Biol. 2013;11(10):e1001695. doi: 10.1371/journal.pbio.1001695. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- JBGS [2021] 118/Seed industry revitalization project of Jiangsu province
- JBGS [2021] 118/Seed industry revitalization project of Jiangsu province
- JBGS [2021] 118/Seed industry revitalization project of Jiangsu province
- JBGS [2021] 118/Seed industry revitalization project of Jiangsu province
- JBGS [2021] 118/Seed industry revitalization project of Jiangsu province
- JBGS [2021] 118/Seed industry revitalization project of Jiangsu province
- JBGS [2021] 118/Seed industry revitalization project of Jiangsu province
- JBGS [2021] 118/Seed industry revitalization project of Jiangsu province
- PZCZ201745/the New cultivar breeding Major Project of Jiangsu province
- PZCZ201745/the New cultivar breeding Major Project of Jiangsu province
- PZCZ201745/the New cultivar breeding Major Project of Jiangsu province
- PZCZ201745/the New cultivar breeding Major Project of Jiangsu province
- PZCZ201745/the New cultivar breeding Major Project of Jiangsu province
- PZCZ201745/the New cultivar breeding Major Project of Jiangsu province
- PZCZ201745/the New cultivar breeding Major Project of Jiangsu province
- PZCZ201745/the New cultivar breeding Major Project of Jiangsu province
- PZCZ201745/the New cultivar breeding Major Project of Jiangsu province
- BK20221207/the Natural Science Foundation of Jiangsu Province
- BK20221207/the Natural Science Foundation of Jiangsu Province
- BK20221207/the Natural Science Foundation of Jiangsu Province
- BK20221207/the Natural Science Foundation of Jiangsu Province
- BK20221207/the Natural Science Foundation of Jiangsu Province
- BK20221207/the Natural Science Foundation of Jiangsu Province
- BK20221207/the Natural Science Foundation of Jiangsu Province
- BK20221207/the Natural Science Foundation of Jiangsu Province
- BK20221207/the Natural Science Foundation of Jiangsu Province
LinkOut - more resources
Full Text Sources

