Localization, traffic and function of Rab34 in adipocyte lipid and endocrine functions
- PMID: 38183057
- PMCID: PMC10770960
- DOI: 10.1186/s12929-023-00990-8
Localization, traffic and function of Rab34 in adipocyte lipid and endocrine functions
Abstract
Background: Excessive lipid accumulation in the adipose tissue in obesity alters the endocrine and energy storage functions of adipocytes. Adipocyte lipid droplets represent key organelles coordinating lipid storage and mobilization in these cells. Recently, we identified the small GTPase, Rab34, in the lipid droplet proteome of adipocytes. Herein, we have characterized the distribution, intracellular transport, and potential contribution of this GTPase to adipocyte physiology and its regulation in obesity.
Methods: 3T3-L1 and human primary preadipocytes were differentiated in vitro and Rab34 distribution and trafficking were analyzed using markers of cellular compartments. 3T3-L1 adipocytes were transfected with expression vectors and/or Rab34 siRNA and assessed for secretory activity, lipid accumulation and expression of proteins regulating lipid metabolism. Proteomic and protein interaction analyses were employed for the identification of the Rab34 interactome. These studies were combined with functional analysis to unveil the role played by the GTPase in adipocytes, with a focus on the actions conveyed by Rab34 interacting proteins. Finally, Rab34 regulation in response to obesity was also evaluated.
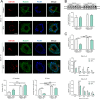
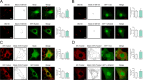
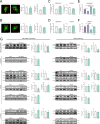
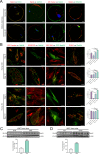
Results: Our results show that Rab34 localizes at the Golgi apparatus in preadipocytes. During lipid droplet biogenesis, Rab34 translocates from the Golgi to endoplasmic reticulum-related compartments and then reaches the surface of adipocyte lipid droplets. Rab34 exerts distinct functions related to its intracellular location. Thus, at the Golgi, Rab34 regulates cisternae integrity as well as adiponectin trafficking and oligomerization. At the lipid droplets, this GTPase controls lipid accumulation and lipolysis through its interaction with the E1-ubiquitin ligase, UBA1, which induces the ubiquitination and proteasomal degradation of the fatty acid transporter and member of Rab34 interactome, FABP5. Finally, Rab34 levels in the adipose tissue and adipocytes are regulated in response to obesity and related pathogenic insults (i.e., fibrosis).
Conclusions: Rab34 plays relevant roles during adipocyte differentiation, including from the regulation of the oligomerization (i.e., biological activity) and secretion of a major adipokine with insulin-sensitizing actions, adiponectin, to lipid storage and mobilization from lipid droplets. Rab34 dysregulation in obesity may contribute to the altered adipokine secretion and lipid metabolism that characterize adipocyte dysfunction in conditions of excess adiposity.
Keywords: Adipocytes; Adiponectin; Golgi apparatus; Lipid droplets; Lipid metabolism; Protein trafficking; Rab34.
© 2024. The Author(s).
Conflict of interest statement
The authors confirm that there are no competing interests.
Figures





Similar articles
-
Rab18 dynamics in adipocytes in relation to lipogenesis, lipolysis and obesity.PLoS One. 2011;6(7):e22931. doi: 10.1371/journal.pone.0022931. Epub 2011 Jul 28. PLoS One. 2011. PMID: 21829560 Free PMC article.
-
Annexin A6 regulates adipocyte lipid storage and adiponectin release.Mol Cell Endocrinol. 2017 Jan 5;439:419-430. doi: 10.1016/j.mce.2016.09.033. Epub 2016 Oct 1. Mol Cell Endocrinol. 2017. PMID: 27702590
-
Peroxiredoxin 3 is a key molecule regulating adipocyte oxidative stress, mitochondrial biogenesis, and adipokine expression.Antioxid Redox Signal. 2012 Feb 1;16(3):229-43. doi: 10.1089/ars.2010.3766. Epub 2011 Oct 17. Antioxid Redox Signal. 2012. PMID: 21902452 Free PMC article.
-
Fat Cell and Fatty Acid Turnover in Obesity.Adv Exp Med Biol. 2017;960:135-160. doi: 10.1007/978-3-319-48382-5_6. Adv Exp Med Biol. 2017. PMID: 28585198 Review.
-
The control of insulin secretion by adipokines: current evidence for adipocyte-beta cell endocrine signalling in metabolic homeostasis.Mamm Genome. 2014 Oct;25(9-10):442-54. doi: 10.1007/s00335-014-9538-7. Epub 2014 Aug 22. Mamm Genome. 2014. PMID: 25146550 Review.
Cited by
-
Gonadal Lipid Storage in Mytilus coruscus: A Comprehensive Gene Network and Key Gene Discovery.Mar Biotechnol (NY). 2025 Jun 19;27(4):99. doi: 10.1007/s10126-025-10475-4. Mar Biotechnol (NY). 2025. PMID: 40536640
-
Moving the fat: Emerging roles of rab GTPases in the regulation of lipid droplet contact sites.Curr Opin Cell Biol. 2025 Apr;93:102466. doi: 10.1016/j.ceb.2025.102466. Epub 2025 Feb 1. Curr Opin Cell Biol. 2025. PMID: 39893800 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

