Enhanced protein secretion in reduced genome strains of Streptomyces lividans
- PMID: 38183102
- PMCID: PMC10768272
- DOI: 10.1186/s12934-023-02269-x
Enhanced protein secretion in reduced genome strains of Streptomyces lividans
Abstract
Background: S. lividans TK24 is a popular host for the production of small molecules and the secretion of heterologous protein. Within its large genome, twenty-nine non-essential clusters direct the biosynthesis of secondary metabolites. We had previously constructed ten chassis strains, carrying deletions in various combinations of specialized metabolites biosynthetic clusters, such as those of the blue actinorhodin (act), the calcium-dependent antibiotic (cda), the undecylprodigiosin (red), the coelimycin A (cpk) and the melanin (mel) clusters, as well as the genes hrdD, encoding a non-essential sigma factor, and matAB, a locus affecting mycelial aggregation. Genome reduction was aimed at reducing carbon flow toward specialized metabolite biosynthesis to optimize the production of secreted heterologous protein.
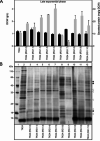
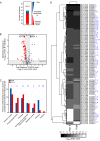
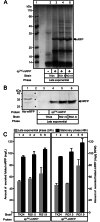
Results: Two of these S. lividans TK24 derived chassis strains showed ~ 15% reduction in biomass yield, 2-fold increase of their total native secretome mass yield and enhanced abundance of several secreted proteins compared to the parental strain. RNAseq and proteomic analysis of the secretome suggested that genome reduction led to cell wall and oxidative stresses and was accompanied by the up-regulation of secretory chaperones and of secDF, a Sec-pathway component. Interestingly, the amount of the secreted heterologous proteins mRFP and mTNFα, by one of these strains, was 12 and 70% higher, respectively, than that secreted by the parental strain.
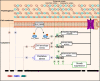
Conclusion: The current study described a strategy to construct chassis strains with enhanced secretory abilities and proposed a model linking the deletion of specialized metabolite biosynthetic clusters to improved production of secreted heterologous proteins.
Keywords: Heterologous secretion; Proteomics; Reduced genome strains; Secretome; Transcriptomics.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Tarkka M, Hampp R. Secondary Metabolites of Soil Streptomycetes in Biotic Interactions. In Secondary Metabolites in Soil Ecology Edited by Karlovsky P. Berlin, Heidelberg: Springer Berlin Heidelberg; 2008: 107–126.[Soil Biology].
-
- Hamed MB, Karamanou S, Olafsdottir S, Basilio JSM, Simoens K, Tsolis KC, Van Mellaert L, Guethmundsdottir EE, Hreggvidsson GO, Anne J, et al. Large-scale production of a thermostable Rhodothermus marinus cellulase by heterologous secretion from Streptomyces lividans. Microb Cell Fact. 2017;16:232. doi: 10.1186/s12934-017-0847-x. - DOI - PMC - PubMed
-
- Lammertyn E, Van Mellaert L, Schacht S, Dillen C, Sablon E, Van Broekhoven A, Anne J. Evaluation of a novel subtilisin inhibitor gene and mutant derivatives for the expression and secretion of mouse Tumor necrosis factor alpha by Streptomyces lividans. Appl Environ Microbiol. 1997;63:1808–13. doi: 10.1128/aem.63.5.1808-1813.1997. - DOI - PMC - PubMed
-
- Pozidis C, Lammertyn E, Politou AS, Anne J, Tsiftsoglou AS, Sianidis G, Economou A. Protein secretion biotechnology using Streptomyces lividans: large-scale production of functional trimeric Tumor necrosis factor alpha. Biotechnol Bioeng. 2001;72:611–9. doi: 10.1002/1097-0290(20010320)72:6<611::AID-BIT1026>3.0.CO;2-0. - DOI - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

