CircPPAP2B controls metastasis of clear cell renal cell carcinoma via HNRNPC-dependent alternative splicing and targeting the miR-182-5p/CYP1B1 axis
- PMID: 38184608
- PMCID: PMC10770969
- DOI: 10.1186/s12943-023-01912-w
CircPPAP2B controls metastasis of clear cell renal cell carcinoma via HNRNPC-dependent alternative splicing and targeting the miR-182-5p/CYP1B1 axis
Abstract
Background: Renal cell carcinoma (RCC) is one of the most common malignant tumor worldwide. Metastasis is a leading case of cancer-related deaths of RCC. Circular RNAs (circRNAs), a class of noncoding RNAs, have emerged as important regulators in cancer metastasis. However, the functional effects and regulatory mechanisms of circRNAs on RCC metastasis remain largely unknown.
Methods: High-throughput RNA sequencing techniques were performed to analyze the expression profiles of circRNAs and mRNAs in highly and poorly invasive clear cell renal cell carcinoma (ccRCC) cell lines. Functional experiments were performed to unveil the regulatory role of circPPAP2B in the proliferation and metastatic capabilities of ccRCC cells. RNA pulldown, Mass spectrometry analysis, RNA methylation immunoprecipitation (MeRIP), RNA immunoprecipitation (RIP), co-immunoprecipitation (CoIP), next-generation RNA-sequencing and double luciferase experiments were employed to clarify the molecular mechanisms by which circPPAP2B promotes ccRCC metastasis.
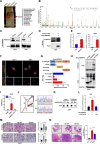
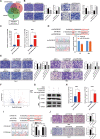
Results: In this study, we describe a newly identified circular RNA called circPPAP2B, which is overexpressed in highly invasive ccRCC cells, as determined through advanced high-throughput RNA sequencing techniques. Furthermore, we observed elevated circPPAP2B in ccRCC tissues, particularly in metastatic ccRCC tissues, and found it to be associated with poor prognosis. Functional experiments unveiled that circPPAP2B actively stimulates the proliferation and metastatic capabilities of ccRCC cells. Mechanistically, circPPAP2B interacts with HNRNPC in a m6A-dependent manner to facilitate HNRNPC nuclear translocation. Subcellular relocalization was dependent upon nondegradable ubiquitination of HNRNPC and stabilization of an HNRNPC/Vimentin/Importin α7 ternary complex. Moreover, we found that circPPAP2B modulates the interaction between HNRNPC and splicing factors, PTBP1 and HNPNPK, and regulates pre-mRNA alternative splicing. Finally, our studies demonstrate that circPPAP2B functions as a miRNA sponge to directly bind to miR-182-5p and increase CYP1B1 expression in ccRCC.
Conclusions: Collectively, our study provides comprehensive evidence that circPPAP2B promotes proliferation and metastasis of ccRCC via HNRNPC-dependent alternative splicing and miR-182-5p/CYP1B1 axis and highlights circPPAP2B as a potential therapeutic target for ccRCC intervention.
Keywords: Alternative splicing; CircPPAP2B; HNRNPC; Nuclear translocation; m6A; miRNA sponge.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Powles T. Recent eUpdate to the ESMO Clinical Practice Guidelines on renal cell carcinoma on cabozantinib and nivolumab for first-line clear cell renal cancer: Renal cell carcinoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2021;32:422–423. doi: 10.1016/j.annonc.2020.11.016. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 82202877/National Natural Science Foundation of China
- 82073162/National Natural Science Foundation of China
- 2023M731561/China Postdoctoral Science Foundation
- 2021C002 and 2020Z005/Present Foundation of Nanfang Hospital, Southern Medical University
- 2021C002 and 2020Z005/Present Foundation of Nanfang Hospital, Southern Medical University
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

