DMT1-dependent endosome-mitochondria interactions regulate mitochondrial iron translocation and metastatic outgrowth
- PMID: 38184712
- PMCID: PMC10890933
- DOI: 10.1038/s41388-023-02933-x
DMT1-dependent endosome-mitochondria interactions regulate mitochondrial iron translocation and metastatic outgrowth
Abstract
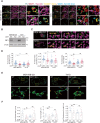
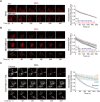
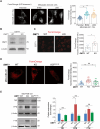
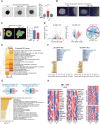
Transient early endosome (EE)-mitochondria interactions can mediate mitochondrial iron translocation, but the associated mechanisms are still elusive. We showed that Divalent Metal Transporter 1 (DMT1) sustains mitochondrial iron translocation via EE-mitochondria interactions in triple-negative MDA-MB-231, but not in luminal A T47D breast cancer cells. DMT1 silencing increases labile iron pool (LIP) levels and activates PINK1/Parkin-dependent mitophagy in MDA-MB-231 cells. Mitochondrial bioenergetics and the iron-associated protein profile were altered by DMT1 silencing and rescued by DMT1 re-expression. Transcriptomic profiles upon DMT1 silencing are strikingly different between 2D and 3D culture conditions, suggesting that the environment context is crucial for the DMT1 knockout phenotype observed in MDA-MB-231 cells. Lastly, in vivo lung metastasis assay revealed that DMT1 silencing promoted the outgrowth of lung metastatic nodules in both human and murine models of triple-negative breast cancer cells. These findings reveal a DMT1-dependent pathway connecting EE-mitochondria interactions to mitochondrial iron translocation and metastatic fitness of breast cancer cells.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Reczek CR, Chandel NS. The two faces of reactive oxygen species in cancer. Annu Rev Cancer Biol. 2017;1:79–98. doi: 10.1146/annurev-cancerbio-041916-065808. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

