Identification of oral therapeutics using an AI platform against the virus responsible for COVID-19, SARS-CoV-2
- PMID: 38186640
- PMCID: PMC10770831
- DOI: 10.3389/fphar.2023.1297924
Identification of oral therapeutics using an AI platform against the virus responsible for COVID-19, SARS-CoV-2
Abstract
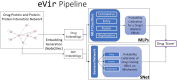
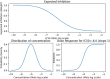
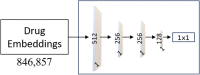
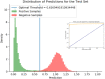
Purpose: This study introduces a sophisticated computational pipeline, eVir, designed for the discovery of antiviral drugs based on their interactions within the human protein network. There is a pressing need for cost-effective therapeutics for infectious diseases (e.g., COVID-19), particularly in resource-limited countries. Therefore, our team devised an Artificial Intelligence (AI) system to explore repurposing opportunities for currently used oral therapies. The eVir system operates by identifying pharmaceutical compounds that mirror the effects of antiviral peptides (AVPs)-fragments of human proteins known to interfere with fundamental phases of the viral life cycle: entry, fusion, and replication. eVir extrapolates the probable antiviral efficacy of a given compound by analyzing its established and predicted impacts on the human protein-protein interaction network. This innovative approach provides a promising platform for drug repurposing against SARS-CoV-2 or any virus for which peptide data is available. Methods: The eVir AI software pipeline processes drug-protein and protein-protein interaction networks generated from open-source datasets. eVir uses Node2Vec, a graph embedding technique, to understand the nuanced connections among drugs and proteins. The embeddings are input a Siamese Network (SNet) and MLPs, each tailored for the specific mechanisms of entry, fusion, and replication, to evaluate the similarity between drugs and AVPs. Scores generated from the SNet and MLPs undergo a Platt probability calibration and are combined into a unified score that gauges the potential antiviral efficacy of a drug. This integrated approach seeks to boost drug identification confidence, offering a potential solution for detecting therapeutic candidates with pronounced antiviral potency. Once identified a number of compounds were tested for efficacy and toxicity in lung carcinoma cells (Calu-3) infected with SARS-CoV-2. A lead compound was further identified to determine its efficacy and toxicity in K18-hACE2 mice infected with SARS-CoV-2. Computational Predictions: The SNet confidently differentiated between similar and dissimilar drug pairs with an accuracy of 97.28% and AUC of 99.47%. Key compounds identified through these networks included Zinc, Mebendazole, Levomenol, Gefitinib, Niclosamide, and Imatinib. Notably, Mebendazole and Zinc showcased the highest similarity scores, while Imatinib, Levemenol, and Gefitinib also ranked within the top 20, suggesting their significant pharmacological potentials. Further examination of protein binding analysis using explainable AI focused on reverse engineering the causality of the networks. Protein interaction scores for Mebendazole and Imatinib revealed their effects on notable proteins such as CDPK1, VEGF2, ABL1, and several tyrosine protein kinases. Laboratory Studies: This study determined that Mebendazole, Gefitinib, Topotecan and to some extent Carfilzomib showed conventional drug-response curves, with IC50 values near or below that of Remdesivir with excellent confidence all above R2>0.91, and no cytotoxicity at the IC50 concentration in Calu-3 cells. Cyclosporine A showed antiviral activity, but also unconventional drug-response curves and low R2 which are explained by the non-dose dependent toxicity of the compound. Additionally, Niclosamide demonstrated a conventional drug-response curve with high confidence; however, its inherent cytotoxicity may be a confounding element that misrepresents true antiviral efficacy, by reflecting cellular damage rather than a genuine antiviral action. Remdesivir was used as a control compound and was evaluated in parallel with the submitted test article and had conventional drug-response curves validating the overall results of the assay. Mebendazole was identified from the cell studies to have efficacy at non-toxic concentrations and were further evaluated in mice infected with SARS-CoV-2. Mebendazole administered to K18-hACE2 mice infected with SARS-CoV-2, resulted in a 44.2% reduction in lung viral load compared to non-treated placebo control respectively. There were no significant differences in body weight and all clinical chemistry determinations evaluated (i.e., kidney and liver enzymes) between the different treatment groups. Conclusion: This research underscores the potential of repurposing existing compounds for treating COVID-19. Our preliminary findings underscore the therapeutic promise of several compounds, notably Mebendazole, in both in vitro and in vivo settings against SARS-CoV-2. Several of the drugs explored, especially Mebendazole, are off-label medication; their cost-effectiveness position them as economical therapies against SARS-CoV-2.
Keywords: COVID-19; SARS-CoV-2; antiviral activity; antiviral peptides; artificial intelligence; infectious diseases; oral therapeutics; siamese networks.
Copyright © 2023 Bess, Berglind, Mukhopadhyay, Brylinski, Alvin, Fattah and Wasan.
Conflict of interest statement
Author KW has stock options in Skymount Medical. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Safety and Efficacy of Imatinib for Hospitalized Adults with COVID-19: A structured summary of a study protocol for a randomised controlled trial.Trials. 2020 Oct 28;21(1):897. doi: 10.1186/s13063-020-04819-9. Trials. 2020. PMID: 33115543 Free PMC article.
-
FDA approved drugs with antiviral activity against SARS-CoV-2: From structure-based repurposing to host-specific mechanisms.Biomed Pharmacother. 2023 Jun;162:114614. doi: 10.1016/j.biopha.2023.114614. Epub 2023 Mar 28. Biomed Pharmacother. 2023. PMID: 37068330 Free PMC article.
-
Current Strategies of Antiviral Drug Discovery for COVID-19.Front Mol Biosci. 2021 May 13;8:671263. doi: 10.3389/fmolb.2021.671263. eCollection 2021. Front Mol Biosci. 2021. PMID: 34055887 Free PMC article. Review.
-
The K18-Human ACE2 Transgenic Mouse Model Recapitulates Non-severe and Severe COVID-19 in Response to an Infectious Dose of the SARS-CoV-2 Virus.J Virol. 2022 Jan 12;96(1):e0096421. doi: 10.1128/JVI.00964-21. Epub 2021 Oct 20. J Virol. 2022. PMID: 34668775 Free PMC article.
-
Current status of antivirals and druggable targets of SARS CoV-2 and other human pathogenic coronaviruses.Drug Resist Updat. 2020 Dec;53:100721. doi: 10.1016/j.drup.2020.100721. Epub 2020 Aug 26. Drug Resist Updat. 2020. PMID: 33132205 Free PMC article. Review.
Cited by
-
cidalsDB: an AI-empowered platform for anti-pathogen therapeutics research.J Cheminform. 2024 Nov 28;16(1):134. doi: 10.1186/s13321-024-00929-7. J Cheminform. 2024. PMID: 39609715 Free PMC article.
References
-
- Abou Dalle I., Kantarjian H., Burger J., Estrov Z., Ohanian M., Verstovsek S., et al. (2019). Efficacy and safety of generic imatinib after switching from original imatinib in patients treated for chronic myeloid leukemia in the United States. Cancer Med. 8, 6559–6565. 10.1002/cam4.2545 - DOI - PMC - PubMed
-
- Assaad H. S., Assaad-Khalil S. (2020). Imatinib a Tyrosine Kinase Inhibitor: a potential treatment for SARS- COV-2 induced pneumonia. Alexandria J. Med. 56 (1), 68–72. 10.1080/20905068.2020.1778417 - DOI
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

