This is a preprint.
Genomic signatures of innovation and selection in the extremotolerant yeast Kluyveromyces marxianus
- PMID: 38187519
- PMCID: PMC10769334
- DOI: 10.1101/2023.12.21.572915
Genomic signatures of innovation and selection in the extremotolerant yeast Kluyveromyces marxianus
Abstract
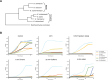
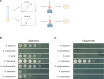
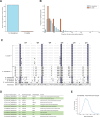
Extremophiles can be the product of millions of years of evolutionary engineering and refinement. The underlying mechanisms can be quite distinct from the ones operating at earlier stages of trait innovation. In this work, we have developed the compost yeast Kluyveromyces marxianus, which diverged from its closest relative >20 million years ago, as a model for interspecies comparative biology and genomics. We applied a battery of growth assays to species of the Kluyveromyces genus and found that K. marxianus outperformed its relatives in a battery of heat and chemical stress conditions. We then generated and analyzed genomes from across the genus, to find derived genetic features associated with, and potentially causal for, K. marxianus traits. We found robust expansions in gene families in the K. marxianus genome, most notably among genes annotated as transmembrane transporters and in metabolism. In molecular-evolution tests, we identified adaptive protein variants at hundreds of genes, among which plasma membrane transporters were over-represented. Together, these signals enable a model for the molecular mechanisms and evolutionary pressures underlying K. marxianus traits, including gains in transporter function mediating stress resistance, and metabolic variants contributing to its capacity for rapid growth in challenging conditions. Such oligogenic architectures may be the rule rather than the exception in phenotypes that have evolved over long timescales.
Figures


References
-
- Al-Qaysi SAS, Abdullah NM, Jaffer MR, Abbas ZA. 2021. Biological Control of Phytopathogenic Fungi by Kluyveromyces marxianus and Torulaspora delbrueckii Isolated from Iraqi Date Vinegar. J Pure Appl Microbiol. 15:300–311.
-
- Am-In S, Yongmanitchai W, Limtong S. 2008. Kluyveromyces siamensis sp. nov., an ascomycetous yeast isolated from water in a mangrove forest in Ranong Province, Thailand. FEMS Yeast Research 8:823–828. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
