This is a preprint.
NOS2 and COX2 Provide Key Spatial Targets that Determine Outcome in ER- Breast Cancer
- PMID: 38187532
- PMCID: PMC10769386
- DOI: 10.1101/2023.12.21.572859
NOS2 and COX2 Provide Key Spatial Targets that Determine Outcome in ER- Breast Cancer
Abstract
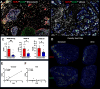
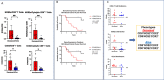
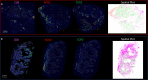
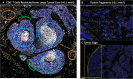
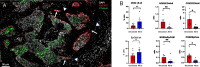
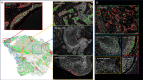
Estrogen receptor-negative (ER-) breast cancer is an aggressive breast cancer subtype with limited therapeutic options. Upregulated expression of both inducible nitric oxide synthase (NOS2) and cyclo-oxygenase (COX2) in breast tumors predicts poor clinical outcomes. Signaling molecules released by these enzymes activate oncogenic pathways, driving cancer stemness, metastasis, and immune suppression. The influence of tumor NOS2/COX2 expression on the landscape of immune markers using multiplex fluorescence imaging of 21 ER- breast tumors were stratified for survival. A powerful relationship between tumor NOS2/COX2 expression and distinct CD8+ T cell phenotypes was observed at 5 years post-diagnosis. These results were confirmed in a validation cohort using gene expression data showing that ratios of NOS2 to CD8 and COX2 to CD8 are strongly associated with poor outcomes in high NOS2/COX2-expressing tumors. Importantly, multiplex imaging identified distinct CD8+ T cell phenotypes relative to tumor NOS2/COX2 expression in Deceased vs Alive patient tumors at 5-year survival. CD8+NOS2-COX2- phenotypes defined fully inflamed tumors with significantly elevated CD8+ T cell infiltration in Alive tumors expressing low NOS2/COX2. In contrast, two distinct phenotypes including inflamed CD8+NOS2+COX2+ regions with stroma-restricted CD8+ T cells and CD8-NOS2-COX2+ immune desert regions with abated CD8+ T cell penetration, were significantly elevated in Deceased tumors with high NOS2/COX2 expression. These results were supported by applying an unsupervised nonlinear dimensionality-reduction technique, UMAP, correlating specific spatial CD8/NOS2/COX2 expression patterns with patient survival. Moreover, spatial analysis of the CD44v6 and EpCAM cancer stem cell (CSC) markers within the CD8/NOS2/COX2 expression landscape revealed positive correlations between EpCAM and inflamed stroma-restricted CD8+NOS2+COX2+ phenotypes at the tumor/stroma interface in deceased patients. Also, positive correlations between CD44v6 and COX2 were identified in immune desert regions in deceased patients. Furthermore, migrating tumor cells were shown to occur only in the CD8-NOS2+COX2+ regions, identifying a metastatic hot spot. Taken together, this study shows the strength of spatial localization analyses of the CD8/NOS2/COX2 landscape, how it shapes the tumor immune microenvironment and the selection of aggressive tumor phenotypes in distinct regions that lead to poor clinical outcomes. This technique could be beneficial for describing tumor niches with increased aggressiveness that may respond to clinically available NOS2/COX2 inhibitors or immune-modulatory agents.
Figures








References
-
- Bauer KR, Brown M, Cress RD, Parise CA, and Caggiano V. Descriptive analysis of estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and HER2-negative invasive breast cancer, the so-called triple-negative phenotype: a population-based study from the California cancer Registry. Cancer. 2007;109(9):1721-8. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
