This is a preprint.
Amyloid precursor protein induces reactive astrogliosis
- PMID: 38187544
- PMCID: PMC10769227
- DOI: 10.1101/2023.12.18.571817
Amyloid precursor protein induces reactive astrogliosis
Update in
-
Amyloid precursor protein induces reactive astrogliosis.Acta Physiol (Oxf). 2024 Jun;240(6):e14142. doi: 10.1111/apha.14142. Epub 2024 Apr 8. Acta Physiol (Oxf). 2024. PMID: 38584589
Abstract
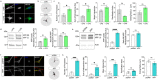
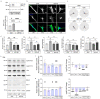
We present in vitro and in vivo evidence demonstrating that Amyloid Precursor Protein (APP) acts as an essential instigator of reactive astrogliosis. Cell-specific overexpression of APP in cultured astrocytes led to remodelling of the intermediate filament network, enhancement of cytokine production and activation of cellular programs centred around the interferon (IFN) pathway, all signs of reactive astrogliosis. Conversely, APP deletion in cultured astrocytes abrogated remodelling of the intermediate filament network and blunted expression of IFN stimulated gene (ISG) products in response to lipopolysaccharide (LPS). Following traumatic brain injury (TBI), mouse reactive astrocytes also exhibited an association between APP and IFN, while APP deletion curbed the increase in glial fibrillary acidic protein (GFAP) observed canonically in astrocytes in response to TBI. Thus, APP represents a molecular inducer and regulator of reactive astrogliosis.
Keywords: amyloid precursor protein; astrocytes; interferon pathway; lipopolysaccharide; reactive astrogliosis; traumatic brain injury.
Figures


References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
