This is a preprint.
Regulation of the transcription factor CdnL promotes adaptation to nutrient stress in Caulobacter
- PMID: 38187569
- PMCID: PMC10769358
- DOI: 10.1101/2023.12.20.572625
Regulation of the transcription factor CdnL promotes adaptation to nutrient stress in Caulobacter
Update in
-
Regulation of the transcription factor CdnL promotes adaptation to nutrient stress in Caulobacter.PNAS Nexus. 2024 Apr 10;3(4):pgae154. doi: 10.1093/pnasnexus/pgae154. eCollection 2024 Apr. PNAS Nexus. 2024. PMID: 38650860 Free PMC article.
Abstract
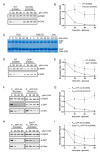
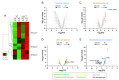
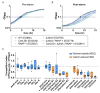
In response to nutrient deprivation, bacteria activate a conserved stress response pathway called the stringent response (SR). During SR activation in Caulobacter crescentus, SpoT synthesizes the secondary messengers (p)ppGpp, which affect transcription by binding RNA polymerase to downregulate anabolic genes. (p)ppGpp also impacts expression of anabolic genes by controlling the levels and activities of their transcriptional regulators. In Caulobacter, a major regulator of anabolic genes is the transcription factor CdnL. If and how CdnL is controlled during the SR and why that might be functionally important is unclear. Here, we show that CdnL is downregulated post-translationally during starvation in a manner dependent on SpoT and the ClpXP protease. Inappropriate stabilization of CdnL during starvation causes misregulation of ribosomal and metabolic genes. Functionally, we demonstrate that the combined action of SR transcriptional regulators and CdnL clearance allows for rapid adaptation to nutrient repletion. Moreover, cells that are unable to clear CdnL during starvation are outcompeted by wild-type cells when subjected to nutrient fluctuations. We hypothesize that clearance of CdnL during the SR, in conjunction with direct binding of (p)ppGpp and DksA to RNAP, is critical for altering the transcriptome in order to permit cell survival during nutrient stress.
Keywords: CarD; CdnL; adaptation; proteolysis; stringent response.
Figures

Similar articles
-
Regulation of the transcription factor CdnL promotes adaptation to nutrient stress in Caulobacter.PNAS Nexus. 2024 Apr 10;3(4):pgae154. doi: 10.1093/pnasnexus/pgae154. eCollection 2024 Apr. PNAS Nexus. 2024. PMID: 38650860 Free PMC article.
-
CdnL, a member of the large CarD-like family of bacterial proteins, is vital for Myxococcus xanthus and differs functionally from the global transcriptional regulator CarD.Nucleic Acids Res. 2010 Aug;38(14):4586-98. doi: 10.1093/nar/gkq214. Epub 2010 Apr 5. Nucleic Acids Res. 2010. PMID: 20371514 Free PMC article.
-
Caulobacter crescentus CdnL is a non-essential RNA polymerase-binding protein whose depletion impairs normal growth and rRNA transcription.Sci Rep. 2017 Feb 24;7:43240. doi: 10.1038/srep43240. Sci Rep. 2017. PMID: 28233804 Free PMC article.
-
House of CarDs: Functional insights into the transcriptional regulator CdnL.Mol Microbiol. 2024 Nov;122(5):789-796. doi: 10.1111/mmi.15268. Epub 2024 Apr 25. Mol Microbiol. 2024. PMID: 38664995 Review.
-
Bacterial lifestyle shapes stringent response activation.Trends Microbiol. 2013 Apr;21(4):174-80. doi: 10.1016/j.tim.2013.01.002. Epub 2013 Feb 16. Trends Microbiol. 2013. PMID: 23419217 Free PMC article. Review.
Cited by
-
Lactylation of Hdac1 regulated by Ldh prevents the pluripotent-to-2C state conversion.Stem Cell Res Ther. 2024 Nov 13;15(1):415. doi: 10.1186/s13287-024-04027-1. Stem Cell Res Ther. 2024. PMID: 39533309 Free PMC article.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
