This is a preprint.
Chromosome fusion and programmed DNA elimination shape karyotypes of parasitic nematodes
- PMID: 38187595
- PMCID: PMC10769430
- DOI: 10.1101/2023.12.21.572835
Chromosome fusion and programmed DNA elimination shape karyotypes of parasitic nematodes
Update in
-
Chromosome fusion and programmed DNA elimination shape karyotypes of nematodes.Curr Biol. 2024 May 20;34(10):2147-2161.e5. doi: 10.1016/j.cub.2024.04.022. Epub 2024 Apr 29. Curr Biol. 2024. PMID: 38688284 Free PMC article.
Abstract
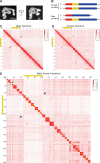
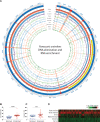
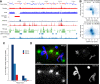
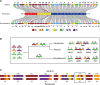
A growing list of metazoans undergo programmed DNA elimination (PDE), where a significant amount of DNA is selectively lost from the somatic genome during development. In some nematodes, PDE leads to the removal and remodeling of the ends of all germline chromosomes. In several species, PDE also generates internal breaks that lead to sequence loss and an increased number of somatic chromosomes. The biological significance of these karyotype changes associated with PDE and the origin and evolution of nematode PDE remain largely unknown. Here, we assembled the single germline chromosome of the horse parasite Parascaris univalens and compared the karyotypes, chromosomal gene organization, and PDE features among ascarid nematodes. We show that PDE in Parascaris converts an XX/XY sex-determination system in the germline into an XX/XO system in the somatic cells. Comparisons of Ascaris, Parascaris, and Baylisascaris ascarid chromosomes suggest that PDE existed in the ancestor of these parasites, and their current distinct germline karyotypes were derived from fusion events of smaller ancestral chromosomes. The DNA breaks involved in PDE resolve these fused germline chromosomes into their pre-fusion karyotypes, leading to alterations in genome architecture and gene expression in the somatic cells. Cytological and genomic analyses further suggest that satellite DNA and the heterochromatic chromosome arms play a dynamic role in the Parascaris germline chromosome during meiosis. Overall, our results show that chromosome fusion and PDE have been harnessed in these ascarids to sculpt their karyotypes, altering the genome organization and serving specific functions in the germline and somatic cells.
Keywords: DNA break; Parascaris; centromere; chromosome fusion; genome; karyotype; meiosis; nematode; programmed DNA elimination; satellite DNA.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


References
-
- Boveri T. (1887). Ueber Differenzierung der Zellkerne wahrend der Furchung des Eies von Ascaris megalocephala. Anat. Anz. 2, 688–693.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
