This is a preprint.
TipN's involvement with centromere segregation in Caulobacter crescentus
- PMID: 38187783
- PMCID: PMC10769339
- DOI: 10.1101/2023.12.20.572679
TipN's involvement with centromere segregation in Caulobacter crescentus
Update in
-
Three factors ParA, TipN, and DnaA-mediated chromosome replication initiation are contributors of centromere segregation in Caulobacter crescentus.Mol Biol Cell. 2024 May 1;35(5):ar68. doi: 10.1091/mbc.E23-12-0503. Epub 2024 Apr 3. Mol Biol Cell. 2024. PMID: 38568781 Free PMC article.
Abstract
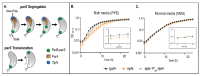
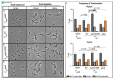
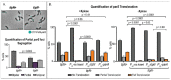
Bacteria's ability to maintain chromosomal integrity throughout their life cycle is crucial for their survival. In Caulobacter crescentus, the polar factor TipN has been proposed to be involved with the partitioning system ParABS. However, cells with tipN knocked out display subtle parS segregation defects. We hypothesized that TipN's role with parS segregation is obscured by other forces that are ParABS-independent. To test our hypothesis, we removed one of those forces - chromosome replication - and analyzed the role of TipN with ParA. We first demonstrate that ParA retains its ability to transport the centromeric region parS from the stalked pole to the opposite pole in the absence of chromosome replication. Our data revealed that in the absence of chromosome replication, TipN becomes essential for ParA's ability to transport parS. Furthermore, we identify a potential connection between the replication initiator DnaA and TipN. Although TipN is not essential for viability, tipN knockout cells lose viability when the regulation of DnaA levels is altered. Our data suggest that the DnaA-dependent susceptibility of tipN knockout cells is connected to parS segregation. Collectively, this work provides insights into the complex regulation involved in the coordination of chromosome replication and segregation in bacteria.
Conflict of interest statement
CONFLICT OF INTEREST The authors declare no conflict of interest.
Figures


Similar articles
-
Three factors ParA, TipN, and DnaA-mediated chromosome replication initiation are contributors of centromere segregation in Caulobacter crescentus.Mol Biol Cell. 2024 May 1;35(5):ar68. doi: 10.1091/mbc.E23-12-0503. Epub 2024 Apr 3. Mol Biol Cell. 2024. PMID: 38568781 Free PMC article.
-
Chromosome Dynamics in Bacteria: Triggering Replication at the Opposite Location and Segregation in the Opposite Direction.mBio. 2019 Jul 30;10(4):e01002-19. doi: 10.1128/mBio.01002-19. mBio. 2019. PMID: 31363028 Free PMC article.
-
ParA's Impact beyond Chromosome Segregation in Caulobacter crescentus.J Bacteriol. 2023 Feb 22;205(2):e0029622. doi: 10.1128/jb.00296-22. Epub 2023 Jan 24. J Bacteriol. 2023. PMID: 36692299 Free PMC article.
-
Bacterial chromosome segregation by the ParABS system.Open Biol. 2020 Jun;10(6):200097. doi: 10.1098/rsob.200097. Epub 2020 Jun 17. Open Biol. 2020. PMID: 32543349 Free PMC article. Review.
-
Multilayered control of chromosome replication in Caulobacter crescentus.Biochem Soc Trans. 2019 Feb 28;47(1):187-196. doi: 10.1042/BST20180460. Epub 2019 Jan 9. Biochem Soc Trans. 2019. PMID: 30626709 Free PMC article. Review.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
