Metagenomic assembly is the main bottleneck in the identification of mobile genetic elements
- PMID: 38188174
- PMCID: PMC10771768
- DOI: 10.7717/peerj.16695
Metagenomic assembly is the main bottleneck in the identification of mobile genetic elements
Abstract
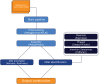
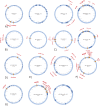
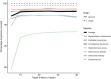
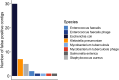
Antimicrobial resistance genes (ARG) are commonly found on acquired mobile genetic elements (MGEs) such as plasmids or transposons. Understanding the spread of resistance genes associated with mobile elements (mARGs) across different hosts and environments requires linking ARGs to the existing mobile reservoir within bacterial communities. However, reconstructing mARGs in metagenomic data from diverse ecosystems poses computational challenges, including genome fragment reconstruction (assembly), high-throughput annotation of MGEs, and identification of their association with ARGs. Recently, several bioinformatics tools have been developed to identify assembled fragments of plasmids, phages, and insertion sequence (IS) elements in metagenomic data. These methods can help in understanding the dissemination of mARGs. To streamline the process of identifying mARGs in multiple samples, we combined these tools in an automated high-throughput open-source pipeline, MetaMobilePicker, that identifies ARGs associated with plasmids, IS elements and phages, starting from short metagenomic sequencing reads. This pipeline was used to identify these three elements on a simplified simulated metagenome dataset, comprising whole genome sequences from seven clinically relevant bacterial species containing 55 ARGs, nine plasmids and five phages. The results demonstrated moderate precision for the identification of plasmids (0.57) and phages (0.71), and moderate sensitivity of identification of IS elements (0.58) and ARGs (0.70). In this study, we aim to assess the main causes of this moderate performance of the MGE prediction tools in a comprehensive manner. We conducted a systematic benchmark, considering metagenomic read coverage, contig length cutoffs and investigating the performance of the classification algorithms. Our analysis revealed that the metagenomic assembly process is the primary bottleneck when linking ARGs to identified MGEs in short-read metagenomics sequencing experiments rather than ARGs and MGEs identification by the different tools.
Keywords: Bacteria; Bacterial; Computational biology; Drug resistance; Metagenomics; Plasmid.
©2024 Kerkvliet et al.
Conflict of interest statement
The authors declare there are no competing interests.
Figures

References
-
- Berbers B, Saltykova A, Garcia-Graells C, Philipp P, Arella F, Marchal K, Winand R, Vanneste K, Roosens NHC, De Keersmaecker SCJ. Combining short and long read sequencing to characterize antimicrobial resistance genes on plasmids applied to an unauthorized genetically modified bacillus. Scientific Reports. 2020;10(1):4310. doi: 10.1038/s41598-020-61158-0. - DOI - PMC - PubMed
-
- Bernard G, Pathmanathan JS, Lannes R, Lopez P, Bapteste E. Microbial dark matter investigations: how microbial studies transform biological knowledge and empirically sketch a logic of scientific discovery. Genome Biology and Evolution. 2018;10(3):707–715. doi: 10.1093/gbe/evy031. - DOI - PMC - PubMed
-
- Bindari YR, Moore RJ, Van TTH, Hilliar M, Wu S-B, Walkden-Brown SW, Gerber PF. Microbial communities of poultry house dust, excreta and litter are partially representative of microbiota of chicken caecum and ileum. PLOS ONE. 2021;16(8):e0255633. doi: 10.1371/journal.pone.0255633. - DOI - PMC - PubMed
-
- Borowiak M, Fischer J, Hammerl JA, Hendriksen RS, Szabo I, Malorny B. Identification of a novel transposon-associated phosphoethanolamine transferase gene, Mcr-5, conferring colistin resistance in d-Tartrate fermenting Salmonella enterica Subsp. Enterica Serovar Paratyphi B. Journal of Antimicrobial Chemotherapy. 2017;72(12):3317–3324. doi: 10.1093/jac/dkx327. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

