Application of artificial scaffold systems in microbial metabolic engineering
- PMID: 38188488
- PMCID: PMC10771841
- DOI: 10.3389/fbioe.2023.1328141
Application of artificial scaffold systems in microbial metabolic engineering
Abstract
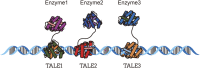
In nature, metabolic pathways are often organized into complex structures such as multienzyme complexes, enzyme molecular scaffolds, or reaction microcompartments. These structures help facilitate multi-step metabolic reactions. However, engineered metabolic pathways in microbial cell factories do not possess inherent metabolic regulatory mechanisms, which can result in metabolic imbalance. Taking inspiration from nature, scientists have successfully developed synthetic scaffolds to enhance the performance of engineered metabolic pathways in microbial cell factories. By recruiting enzymes, synthetic scaffolds facilitate the formation of multi-enzyme complexes, leading to the modulation of enzyme spatial distribution, increased enzyme activity, and a reduction in the loss of intermediate products and the toxicity associated with harmful intermediates within cells. In recent years, scaffolds based on proteins, nucleic acids, and various organelles have been developed and employed to facilitate multiple metabolic pathways. Despite varying degrees of success, synthetic scaffolds still encounter numerous challenges. The objective of this review is to provide a comprehensive introduction to these synthetic scaffolds and discuss their latest research advancements and challenges.
Keywords: enzyme molecular scaffolds; microbial cell factory; multienzyme complexes; reaction microcompartments; synthetic scaffold.
Copyright © 2023 Liu, Dong, Yang, Li and Chiu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Synthetic Scaffold Systems for Increasing the Efficiency of Metabolic Pathways in Microorganisms.Biology (Basel). 2021 Mar 11;10(3):216. doi: 10.3390/biology10030216. Biology (Basel). 2021. PMID: 33799683 Free PMC article. Review.
-
[Research progress on synthetic scaffold in metabolic engineering - a review].Sheng Wu Gong Cheng Xue Bao. 2019 Mar 25;35(3):363-374. doi: 10.13345/j.cjb.180298. Sheng Wu Gong Cheng Xue Bao. 2019. PMID: 30912345 Review. Chinese.
-
Mechanistic Aspects for the Modulation of Enzyme Reactions on the DNA Scaffold.Molecules. 2022 Sep 24;27(19):6309. doi: 10.3390/molecules27196309. Molecules. 2022. PMID: 36234845 Free PMC article. Review.
-
Synthetic scaffolds for pathway enhancement.Curr Opin Biotechnol. 2015 Dec;36:98-106. doi: 10.1016/j.copbio.2015.08.009. Epub 2015 Aug 29. Curr Opin Biotechnol. 2015. PMID: 26322735 Review.
-
Synthetic protein scaffolds for the colocalisation of co-acting enzymes.Biotechnol Adv. 2020 Nov 15;44:107627. doi: 10.1016/j.biotechadv.2020.107627. Epub 2020 Aug 29. Biotechnol Adv. 2020. PMID: 32871185 Review.
Cited by
-
Biosynthesis of Edible Terpenoids: Hosts and Applications.Foods. 2025 Feb 17;14(4):673. doi: 10.3390/foods14040673. Foods. 2025. PMID: 40002116 Free PMC article. Review.
References
Publication types
LinkOut - more resources
Full Text Sources

