Diverse bacteria elicit distinct neutrophil responses in a physiologically relevant model of infection
- PMID: 38188520
- PMCID: PMC10770534
- DOI: 10.1016/j.isci.2023.108627
Diverse bacteria elicit distinct neutrophil responses in a physiologically relevant model of infection
Abstract
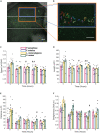
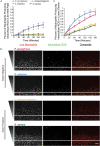
An efficient neutrophil response is critical for fighting bacterial infections, which remain a significant global health concern; therefore, modulating neutrophil function could be an effective therapeutic approach. While we have a general understanding of how neutrophils respond to bacteria, how neutrophil function differs in response to diverse bacterial infections remains unclear. Here, we use a microfluidic infection-on-a-chip device to investigate the neutrophil response to four bacterial species: Pseudomonas aeruginosa, Salmonella enterica, Listeria monocytogenes, and Staphylococcus aureus. We find enhanced neutrophil extravasation to L. monocytogenes, a limited overall response to S. aureus, and identify IL-6 as universally important for neutrophil extravasation. Furthermore, we demonstrate a higher percentage of neutrophils generate reactive oxygen species (ROS) when combating gram-negative bacteria versus gram-positive bacteria. For all bacterial species, we found the percentage of neutrophils producing ROS increased following extravasation through an endothelium, underscoring the importance of studying neutrophil function in physiologically relevant models.
Keywords: Bacteriology; Immunology.
© 2023 The Author(s).
Conflict of interest statement
None to declare.
Figures





Similar articles
-
Collagen concentration regulates neutrophil extravasation and migration in response to infection in an endothelium dependent manner.Front Immunol. 2024 Jul 3;15:1405364. doi: 10.3389/fimmu.2024.1405364. eCollection 2024. Front Immunol. 2024. PMID: 39021568 Free PMC article.
-
CD137 expressed on neutrophils plays dual roles in antibacterial responses against Gram-positive and Gram-negative bacterial infections.Infect Immun. 2013 Jun;81(6):2168-77. doi: 10.1128/IAI.00115-13. Epub 2013 Apr 1. Infect Immun. 2013. PMID: 23545301 Free PMC article.
-
NLRP6 negatively regulates pulmonary host defense in Gram-positive bacterial infection through modulating neutrophil recruitment and function.PLoS Pathog. 2018 Sep 24;14(9):e1007308. doi: 10.1371/journal.ppat.1007308. eCollection 2018 Sep. PLoS Pathog. 2018. PMID: 30248149 Free PMC article.
-
Epic Immune Battles of History: Neutrophils vs. Staphylococcus aureus.Front Cell Infect Microbiol. 2017 Jun 30;7:286. doi: 10.3389/fcimb.2017.00286. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 28713774 Free PMC article. Review.
-
Sulopenem: An Intravenous and Oral Penem for the Treatment of Urinary Tract Infections Due to Multidrug-Resistant Bacteria.Drugs. 2022 Apr;82(5):533-557. doi: 10.1007/s40265-022-01688-1. Epub 2022 Mar 16. Drugs. 2022. PMID: 35294769 Review.
Cited by
-
Gut microbiota deficiency reduces neutrophil activation and is protective after ischemic stroke.J Neuroinflammation. 2025 May 23;22(1):137. doi: 10.1186/s12974-025-03448-w. J Neuroinflammation. 2025. PMID: 40410847 Free PMC article.
-
Pleural fluid proteomics from patients with pleural infection shows signatures of diverse neutrophilic responses: The Oxford Pleural Infection Endotyping Study (TORPIDS-2).Eur Respir J. 2025 Jul 14;66(1):2500010. doi: 10.1183/13993003.00010-2025. Print 2025 Jul. Eur Respir J. 2025. PMID: 40246538 Free PMC article.
-
Collagen concentration regulates neutrophil extravasation and migration in response to infection in an endothelium dependent manner.Front Immunol. 2024 Jul 3;15:1405364. doi: 10.3389/fimmu.2024.1405364. eCollection 2024. Front Immunol. 2024. PMID: 39021568 Free PMC article.
-
Correlation of Neutrophil-Lymphocyte Ratio and Critical Illness in Adults on Vancomycin: A Cross-Sectional Study.Int J Gen Med. 2025 Jul 31;18:4157-4167. doi: 10.2147/IJGM.S537039. eCollection 2025. Int J Gen Med. 2025. PMID: 40766859 Free PMC article.
-
THP-1 Macrophages Limit Neutrophil Transendothelial Migration in a Model Infection.Cell Mol Bioeng. 2024 Jul 20;17(4):279-293. doi: 10.1007/s12195-024-00813-2. eCollection 2024 Aug. Cell Mol Bioeng. 2024. PMID: 39372553 Free PMC article.
References
-
- Moreland J.G., Bailey G., Nauseef W.M., Weiss J.P. Organism-Specific Neutrophil-Endothelial Cell Interactions in Response to Escherichia coli , Streptococcus pneumoniae , and Staphylococcus aureus. J. Immunol. 2004;172:426–432. - PubMed
-
- Angus D.C., van der Poll T. Severe Sepsis and Septic Shock. N. Engl. J. Med. 2013;369:840–851. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

