The metaproteome of the gut microbiota in pediatric patients affected by COVID-19
- PMID: 38188629
- PMCID: PMC10766818
- DOI: 10.3389/fcimb.2023.1327889
The metaproteome of the gut microbiota in pediatric patients affected by COVID-19
Abstract
Introduction: The gut microbiota (GM) play a significant role in the infectivity and severity of COVID-19 infection. However, the available literature primarily focuses on adult patients and it is known that the microbiota undergoes changes throughout the lifespan, with significant alterations occurring during infancy and subsequently stabilizing during adulthood. Moreover, children have exhibited milder symptoms of COVID-19 disease, which has been associated with the abundance of certain protective bacteria. Here, we examine the metaproteome of pediatric patients to uncover the biological mechanisms that underlie this protective effect of the GM.
Methods: We performed nanoliquid chromatography coupled with tandem mass spectrometry on a high resolution analytical platform, resulting in label free quantification of bacterial protein groups (PGs), along with functional annotations via COG and KEGG databases by MetaLab-MAG. Additionally, taxonomic assignment was possible through the use of the lowest common ancestor algorithm provided by Unipept software.
Results: A COVID-19 GM functional dissimilarity respect to healthy subjects was identified by univariate analysis. The alteration in COVID-19 GM function is primarily based on bacterial pathways that predominantly involve metabolic processes, such as those related to tryptophan, butanoate, fatty acid, and bile acid biosynthesis, as well as antibiotic resistance and virulence.
Discussion: These findings highlight the mechanisms by which the pediatric GM could contribute to protection against the more severe manifestations of the disease in children. Uncovering these mechanisms can, therefore, have important implications in the discovery of novel adjuvant therapies for severe COVID-19.
Keywords: COVID-19; SARS-CoV-2; functional patterns; gut microbiota; mass spectrometry; metaproteomics; pediatrics.
Copyright © 2023 Marzano, Mortera, Marangelo, Piazzesi, Rapisarda, Pane, Del Chierico, Vernocchi, Romani, Campana, Palma, Putignani and the CACTUS Study Team.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures


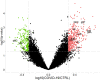


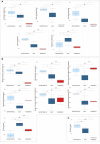

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

