The complete mitochondrial genome of deep-sea ophiuroid Ophioleila elegans (Echinodermata: Ophiuroidea) from the Shkolnik Guyot, a northwest Pacific seamount
- PMID: 38189027
- PMCID: PMC10768936
- DOI: 10.1080/23802359.2023.2288441
The complete mitochondrial genome of deep-sea ophiuroid Ophioleila elegans (Echinodermata: Ophiuroidea) from the Shkolnik Guyot, a northwest Pacific seamount
Abstract
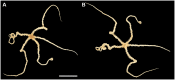
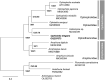
Ophiuroids are a diversified benthic taxon in the deep sea. Given their various dispersal strategies, they are considered an adequate group to assess genetic connectivity, especially in the seamounts that function as islands. Ophioleila elegans A.H. Clark, 1949, in the family Ophiothamnidae, was previously reported from the Caiwei Guyot, a seamount in the northwest Pacific Ocean. Here, we described the mitochondrial genome of O. elegans collected from another seamount in the northwest Pacific. The whole mitogenome is 16,376 bp in length and encodes 13 protein-coding genes, two ribosomal RNA genes, and 22 transfer RNA genes. Phylogenetic analysis based on the mitogenome sequences showed that O. elegans was clustered with Histampica sp., the only species for which mitogenome sequence has been reported within the family Ophiothamnidae. The complete mitogenome of O. elegans first reported in the present study provides useful information for population genetics and evolutionary relationship of this taxon, especially in the northwest Pacific seamounts.
Keywords: Deep-sea Ophiuroidea; Ophioleila elegans; mitochondrial genome; phylogeny; seamount.
© 2023 The Author(s). Published by Informa UK Limited, trading as Taylor & Francis Group.
Conflict of interest statement
The authors report that there are no conflicts of interest.
Figures

References
-
- Cho W, Shank TM.. 2010. Incongruent patterns of genetic connectivity among four ophiuroid species with differing coral host specificity on North Atlantic seamounts. Mar Ecol. 31(Suppl. 1):121–143. doi: 10.1111/j.1439-0485.2010.00395.x. - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
