Candidatus Methanosphaera massiliense sp. nov., a methanogenic archaeal species found in a human fecal sample and prevalent in pigs and red kangaroos
- PMID: 38189277
- PMCID: PMC10845953
- DOI: 10.1128/spectrum.05141-22
Candidatus Methanosphaera massiliense sp. nov., a methanogenic archaeal species found in a human fecal sample and prevalent in pigs and red kangaroos
Abstract
Methanosphaera stadtmanae was the sole Methanosphaera representative to be cultured and detected by molecular methods in the human gut microbiota, further associated with digestive and respiratory diseases, leaving unknown the actual diversity of human-associated Methanosphaera species. Here, a novel Methanosphaera species, Candidatus Methanosphaera massiliense (Ca. M. massiliense) sp. nov. was isolated by culture using a hydrogen- and carbon dioxide-free medium from one human feces sample. Ca. M. massiliense is a non-motile, 850 nm Gram-positive coccus autofluorescent at 420 nm. Whole-genome sequencing yielded a 29.7% GC content, gapless 1,785,773 bp genome sequence with an 84.5% coding ratio, encoding for alcohol and aldehyde dehydrogenases promoting the growth of Ca. M. massiliense without hydrogen. Screening additional mammal and human feces using a specific genome sequence-derived DNA-polymerase RT-PCR system yielded a prevalence of 22% in pigs, 12% in red kangaroos, and no detection in 149 other human samples. This study, extending the diversity of Methanosphaera in human microbiota, questions the zoonotic sources of Ca. M. massiliense and possible transfer between hosts.IMPORTANCEMethanogens are constant inhabitants in the human gut microbiota in which Methanosphaera stadtmanae was the only cultivated Methanosphaera representative. We grew Candidatus Methanosphaera massiliense sp. nov. from one human feces sample in a novel culture medium under a nitrogen atmosphere. Systematic research for methanogens in human and animal fecal samples detected Ca. M. massiliense in pig and red kangaroo feces, raising the possibility of its zoonotic acquisition. Host specificity, source of acquisition, and adaptation of methanogens should be further investigated.
Keywords: Candidatus Methanosphaera massiliense; Methanosphaera; host transfer; human gut microbiota; hydrogen-free culture; methanogens.
Conflict of interest statement
V.P., M.D., G.A., G.G., and E.T. are co-inventors of a patented culture medium referred to as GG culture medium in this report (patent FR 23 01404).
Figures

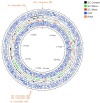
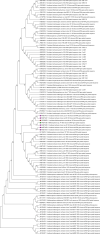

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

