Bacterial pathogens in Xpert MTB/RIF Ultra-negative sputum samples of patients with presumptive tuberculosis in a high TB burden setting: a 16S rRNA analysis
- PMID: 38189296
- PMCID: PMC10845949
- DOI: 10.1128/spectrum.02931-23
Bacterial pathogens in Xpert MTB/RIF Ultra-negative sputum samples of patients with presumptive tuberculosis in a high TB burden setting: a 16S rRNA analysis
Abstract
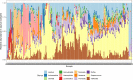
In patients with presumptive tuberculosis (TB) in whom the diagnosis of TB was excluded, understanding the bacterial etiology of lower respiratory tract infections (LRTIs) is important for optimal patient management. A secondary analysis was performed on a cohort of 250 hospitalized patients with symptoms of TB. Bacterial DNA was extracted from sputum samples for Illumina 16S rRNA sequencing to identify bacterial species based on amplicon sequence variant level. The bacterial pathogen most likely to be responsible for the patients' LRTI could only be identified in a minority (6.0%, 13/215) of cases based on 16S rRNA amplicon sequencing: Mycoplasma pneumoniae (n = 7), Bordetella pertussis (n = 2), Acinetobacter baumanii (n = 2), and Pseudomonas aeruginosa (n = 2). Other putative pathogens were present in similar proportions of Xpert Ultra-positive and Xpert Ultra-negative sputum samples. The presence of Streptococcus (pseudo)pneumoniae appeared to increase the odds of radiological abnormalities (aOR 2.5, 95% CI 1.12-6.16) and the presence of S. (pseudo)pneumoniae (aOR 5.31, 95% CI 1.29-26.6) and Moraxella catarrhalis/nonliquefaciens (aOR 12.1, 95% CI 2.67-72.8) increased the odds of 6-month mortality, suggesting that these pathogens might have clinical relevance. M. pneumoniae, B. pertussis, and A. baumanii appeared to be the possible causes of TB-like symptoms. S. (pseudo)pneumoniae and M. catarrhalis/nonliquefaciens also appeared of clinical relevance based on 16S rRNA amplicon sequencing. Further research using tools with higher discriminatory power than 16S rRNA sequencing is required to develop optimal diagnostic and treatment strategies for this population.IMPORTANCEThe objective of this study was to identify possible bacterial lower respiratory tract infection (LRTI) pathogens in hospitalized patients who were initially suspected to have TB but later tested negative using the Xpert Ultra test. Although 16S rRNA was able to identify some less common or difficult-to-culture pathogens such as Mycoplasma pneumoniae and Bordetella pertussis, one of the main findings of the study is that, in contrast to what we had hypothesized, 16S rRNA is not a method that can be used to assist in the management of patients with presumptive TB having a negative Xpert Ultra test. Even though this could be considered a negative finding, we believe it is an important finding to report as it highlights the need for further research using different approaches.
Keywords: Ethiopia; LRTIs; M. tuberculosis; bacterial etiology; diagnostics; presumptive TB cases; sequencing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
The New Xpert MTB/RIF Ultra: Improving Detection of Mycobacterium tuberculosis and Resistance to Rifampin in an Assay Suitable for Point-of-Care Testing.mBio. 2017 Aug 29;8(4):e00812-17. doi: 10.1128/mBio.00812-17. mBio. 2017. PMID: 28851844 Free PMC article.
-
Xpert MTB/RIF Ultra and Xpert MTB/RIF for diagnosis of tuberculosis in an HIV-endemic setting with a high burden of previous tuberculosis: a two-cohort diagnostic accuracy study.Lancet Respir Med. 2020 Apr;8(4):368-382. doi: 10.1016/S2213-2600(19)30370-4. Epub 2020 Feb 14. Lancet Respir Med. 2020. PMID: 32066534
-
Evaluation of Xpert MTB/RIF Ultra performance for pulmonary tuberculosis diagnosis on smear-negative respiratory samples in a French centre.Eur J Clin Microbiol Infect Dis. 2019 Mar;38(3):601-605. doi: 10.1007/s10096-018-03463-1. Epub 2019 Jan 24. Eur J Clin Microbiol Infect Dis. 2019. PMID: 30680567
-
Xpert MTB/RIF and Xpert Ultra assays for screening for pulmonary tuberculosis and rifampicin resistance in adults, irrespective of signs or symptoms.Cochrane Database Syst Rev. 2021 Mar 23;3(3):CD013694. doi: 10.1002/14651858.CD013694.pub2. Cochrane Database Syst Rev. 2021. PMID: 33755189 Free PMC article.
-
Diagnostic accuracy of the new Xpert MTB/RIF Ultra for tuberculosis disease: A preliminary systematic review and meta-analysis.Int J Infect Dis. 2020 Jan;90:35-45. doi: 10.1016/j.ijid.2019.09.016. Epub 2019 Sep 20. Int J Infect Dis. 2020. PMID: 31546008
Cited by
-
Coinfections in Tuberculosis in Low- and Middle-Income Countries: Epidemiology, Clinical Implications, Diagnostic Challenges, and Management Strategies-A Narrative Review.J Clin Med. 2025 Mar 21;14(7):2154. doi: 10.3390/jcm14072154. J Clin Med. 2025. PMID: 40217604 Free PMC article. Review.
References
-
- Yigezu A, Misganaw A, Getnet F, Berheto TM, Walker A, Zergaw A, Gobena FA, Haile MA, Hailu A, Memirie ST, et al. 2023. Burden of lower respiratory infections and associated risk factors across regions in Ethiopia: a subnational analysis of the Global Burden of Diseases 2019 study. BMJ Open 13:e068498. doi: 10.1136/bmjopen-2022-068498 - DOI - PMC - PubMed
-
- Dasaraju PV CL. 1996. Infections of the respiratory system. In Baron S (ed), (ed), Medical microbiology. University of Texas Medical Branch at Galveston, Galveston. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical