CPSF3 inhibition blocks pancreatic cancer cell proliferation through disruption of core histone mRNA processing
- PMID: 38191171
- PMCID: PMC10870380
- DOI: 10.1261/rna.079931.123
CPSF3 inhibition blocks pancreatic cancer cell proliferation through disruption of core histone mRNA processing
Abstract
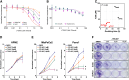
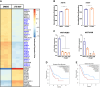
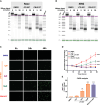
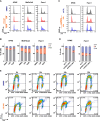
Pancreatic ductal adenocarcinoma (PDAC) is a lethal disease with limited effective treatment options, potentiating the importance of uncovering novel drug targets. Here, we target cleavage and polyadenylation specificity factor 3 (CPSF3), the 3' endonuclease that catalyzes mRNA cleavage during polyadenylation and histone mRNA processing. We find that CPSF3 is highly expressed in PDAC and is associated with poor prognosis. CPSF3 knockdown blocks PDAC cell proliferation and colony formation in vitro and tumor growth in vivo. Chemical inhibition of CPSF3 by the small molecule JTE-607 also attenuates PDAC cell proliferation and colony formation, while it has no effect on cell proliferation of nontransformed immortalized control pancreatic cells. Mechanistically, JTE-607 induces transcriptional readthrough in replication-dependent histones, reduces core histone expression, destabilizes chromatin structure, and arrests cells in the S-phase of the cell cycle. Therefore, CPSF3 represents a potential therapeutic target for the treatment of PDAC.
Keywords: CPSF3; JTE-607; alternative polyadenylation; chromatin stability; histone processing.
© 2024 Alahmari et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures



References
-
- Arnadottir GA, Oddsson A, Jensson BO, Gisladottir S, Simon MT, Arnthorsson AO, Katrinardottir H, Fridriksdottir R, Ivarsdottir EV, Jonasdottir A, et al. 2022. Population-level deficit of homozygosity unveils CPSF3 as an intellectual disability syndrome gene. Nat Commun 13: 705. 10.1038/s41467-022-28330-8 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
