A cyclin D1 intrinsically disordered domain accesses modified histone motifs to govern gene transcription
- PMID: 38191593
- PMCID: PMC10774418
- DOI: 10.1038/s41389-023-00502-1
A cyclin D1 intrinsically disordered domain accesses modified histone motifs to govern gene transcription
Abstract
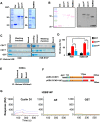
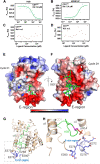
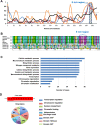
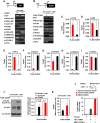
The essential G1-cyclin, CCND1, is frequently overexpressed in cancer, contributing to tumorigenesis by driving cell-cycle progression. D-type cyclins are rate-limiting regulators of G1-S progression in mammalian cells via their ability to bind and activate CDK4 and CDK6. In addition, cyclin D1 conveys kinase-independent transcriptional functions of cyclin D1. Here we report that cyclin D1 associates with H2BS14 via an intrinsically disordered domain (IDD). The same region of cyclin D1 was necessary for the induction of aneuploidy, induction of the DNA damage response, cyclin D1-mediated recruitment into chromatin, and CIN gene transcription. In response to DNA damage H2BS14 phosphorylation occurs, resulting in co-localization with γH2AX in DNA damage foci. Cyclin D1 ChIP seq and γH2AX ChIP seq revealed ~14% overlap. As the cyclin D1 IDD functioned independently of the CDK activity to drive CIN, the IDD domain may provide a rationale new target to complement CDK-extinction strategies.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

