A palmitoyl transferase chemical-genetic system to map ZDHHC-specific S-acylation
- PMID: 38191663
- PMCID: PMC11471619
- DOI: 10.1038/s41587-023-02030-0
A palmitoyl transferase chemical-genetic system to map ZDHHC-specific S-acylation
Abstract
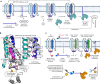
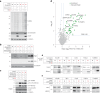
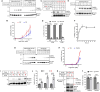
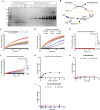
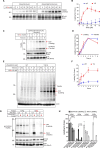
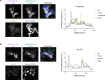
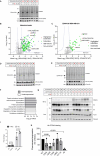
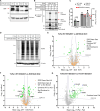
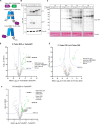
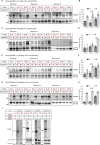
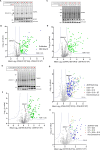
The 23 human zinc finger Asp-His-His-Cys motif-containing (ZDHHC) S-acyltransferases catalyze long-chain S-acylation at cysteine residues across an extensive network of hundreds of proteins important for normal physiology or dysregulated in disease. Here we present a technology to directly map the protein substrates of a specific ZDHHC at the whole-proteome level, in intact cells. Structure-guided engineering of paired ZDHHC 'hole' mutants and 'bumped' chemically tagged fatty acid probes enabled probe transfer to specific protein substrates with excellent selectivity over wild-type ZDHHCs. Chemical-genetic systems were exemplified for five human ZDHHCs (3, 7, 11, 15 and 20) and applied to generate de novo ZDHHC substrate profiles, identifying >300 substrates and S-acylation sites for new functionally diverse proteins across multiple cell lines. We expect that this platform will elucidate S-acylation biology for a wide range of models and organisms.
© 2024. The Author(s).
Conflict of interest statement
E.W.T. is a founder and shareholder in Myricx Pharma and receives consultancy or research funding from Kura Oncology, Pfizer, Samsara Therapeutics, Myricx Pharma, Merck Sharp and Dohme (MSD), Exscientia and Daiichi Sankyo. J.D. has acted as a consultant for AstraZeneca, Jubilant, Theras, BridgeBio and Vividion and receives research funding from Bristol Myers Squibb and Revolution Medicines. The other authors declare no competing interests.
Figures





References
-
- Lanyon-Hogg, T., Faronato, M., Serwa, R. A. & Tate, E. W. Dynamic protein acylation: new substrates, mechanisms, and drug targets. Trends Biochem. Sci.42, 566–581 (2017). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

