Depletion of JunB increases adipocyte thermogenic capacity and ameliorates diet-induced insulin resistance
- PMID: 38191667
- PMCID: PMC10954369
- DOI: 10.1038/s42255-023-00945-1
Depletion of JunB increases adipocyte thermogenic capacity and ameliorates diet-induced insulin resistance
Abstract
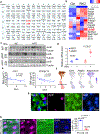
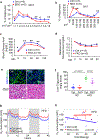
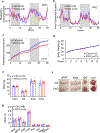
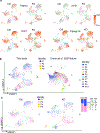
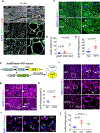
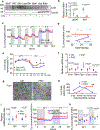
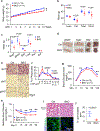
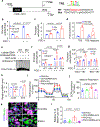
The coexistence of brown adipocytes with low and high thermogenic activity is a fundamental feature of brown adipose tissue heterogeneity and plasticity. However, the mechanisms that govern thermogenic adipocyte heterogeneity and its significance in obesity and metabolic disease remain poorly understood. Here we show that in male mice, a population of transcription factor jun-B (JunB)-enriched (JunB+) adipocytes within the brown adipose tissue exhibits lower thermogenic capacity compared to high-thermogenic adipocytes. The JunB+ adipocyte population expands in obesity. Depletion of JunB in adipocytes increases the fraction of adipocytes exhibiting high thermogenic capacity, leading to enhanced basal and cold-induced energy expenditure and protection against diet-induced obesity and insulin resistance. Mechanistically, JunB antagonizes the stimulatory effects of PPARγ coactivator-1α on high-thermogenic adipocyte formation by directly binding to the promoter of oestrogen-related receptor alpha, a PPARγ coactivator-1α downstream effector. Taken together, our study uncovers that JunB shapes thermogenic adipocyte heterogeneity, serving a critical role in maintaining systemic metabolic health.
© 2024. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures








References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

