Validated assays for the quantification of C9orf72 human pathology
- PMID: 38191789
- PMCID: PMC10774390
- DOI: 10.1038/s41598-023-50667-3
Validated assays for the quantification of C9orf72 human pathology
Abstract
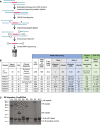
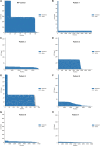
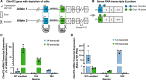
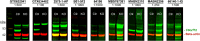
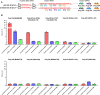
A repeat expansion mutation in the C9orf72 gene is the leading known genetic cause of FTD and ALS. The C9orf72-ALS/FTD field has been plagued by a lack of reliable tools to monitor this genomic locus and its RNA and protein products. We have validated assays that quantify C9orf72 pathobiology at the DNA, RNA and protein levels using knock-out human iPSC lines as controls. Here we show that single-molecule sequencing can accurately measure the repeat expansion and faithfully report on changes to the C9orf72 locus in what has been a traditionally hard to sequence genomic region. This is of particular value to sizing and phasing the repeat expansion and determining changes to the gene locus after gene editing. We developed ddPCR assays to quantify two major C9orf72 transcript variants, which we validated by selective excision of their distinct transcriptional start sites. Using validated knock-out human iPSC lines, we validated 4 commercially available antibodies (of 9 tested) that were specific for C9orf72 protein quantification by Western blot, but none were specific for immunocytochemistry. We tested 15 combinations of antibodies against dipeptide repeat proteins (DPRs) across 66 concentrations using MSD immunoassay, and found two (against poly-GA and poly-GP) that yielded a 1.5-fold or greater signal increase in patient iPSC-motor neurons compared to knock-out control, and validated them in human postmortem and transgenic mouse brain tissue. Our validated DNA, RNA and protein assays are applicable to discovery research as well as clinical trials.
© 2024. The Author(s).
Conflict of interest statement
B.R.C. is a founder of Tenaya Therapeutics (
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

