Whole-exome sequencing of Nigerian benign prostatic hyperplasia reveals increased alterations in apoptotic pathways
- PMID: 38192023
- PMCID: PMC10922327
- DOI: 10.1002/pros.24662
Whole-exome sequencing of Nigerian benign prostatic hyperplasia reveals increased alterations in apoptotic pathways
Abstract
Background: Through whole-exome sequencing of 60 formalin-fixed paraffin-embedded Nigerian (NGRn) benign prostatic hyperplasia (BPH) samples, we identified germline and somatic alterations in apoptotic pathways impacting BPH development and progression. Prostate enlargement is a common occurrence in male aging; however, this enlargement can lead to lower urinary tract symptoms that negatively impact quality of life. This impact is disproportionately present in men of African ancestry. BPH pathophysiology is poorly understood and studies examining non-European populations are lacking.
Methods: In this study, NGRn BPH, normal prostate, and prostate cancer (PCa) tumor samples were sequenced and compared to characterize genetic alterations in NGRn BPH.
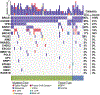
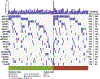
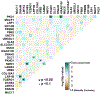
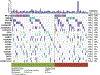
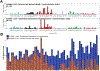
Results: Two hundred and two nonbenign, ClinVar-annotated germline variants were present in NGRn BPH samples. Six genes [BRCA1 (92%), HSD3B1 (85%), TP53 (37%), PMS2 (23%), BARD1 (20%), and BRCA2 (17%)] were altered in at least 10% of samples; however, compared to NGRn normal and tumor, the frequency of alterations in BPH samples showed no significant differences at the gene or variant level. BRCA2_rs11571831 and TP53_rs1042522 germline alterations had a statistically significant co-occurrence interaction in BPH samples. In at least two BPH samples, 173 genes harbored somatic variants known to be clinically actionable. Three genes (COL18A1, KIF16B, and LRP1) showed a statistically significant (p < 0.05) higher frequency in BPH. NGRn BPH also had five gene pairs (PKD1/KIAA0100, PKHD1/PKD1, DNAH9/LRP1B, NWD1/DCHS2, and TCERG1/LMTK2) with statistically significant co-occurring interactions. Two hundred and seventy-nine genes contained novel somatic variants in NGRn BPH. Three genes (CABP1, FKBP1C, and RP11-595B24.2) had a statistically significant (p < 0.05) higher alteration frequency in NGRn BPH and three were significantly higher in NGRn tumor (CACNA1A, DMKN, and CACNA2D2). Pairwise Fisher's exact tests showed 14 gene pairs with statistically significant (p < 0.05) interactions and four interactions approaching significance (p < 0.10). Mutational patterns in NGRn BPH were similar to COSMIC (Catalog of Somatic Mutations in Cancer) signatures associated with aging and dysfunctional DNA damage repair.
Conclusions: NGRn BPH contained significant germline alteration interactions (BRCA2_rs11571831 and TP53_rs1042522) and increased somatic alteration frequencies (LMTK2, LRP1, COL18A1, CABP1, and FKBP1C) that impact apoptosis. Normal prostate development is maintained by balancing apoptotic and proliferative activity. Dysfunction in either mechanism can lead to abnormal prostate growth. This work is the first to examine genomic sequencing in NGRn BPH and provides data that fill known gaps in the understanding BPH and how it impacts men of African ancestry.
Keywords: African Ancestry; apoptosis; benign prostatic hyperplasia; germline variants; somatic variants; whole-exome sequencing.
© 2024 Wiley Periodicals LLC.
Figures

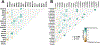

Similar articles
-
Whole-exome Sequencing of Nigerian Prostate Tumors from the Prostate Cancer Transatlantic Consortium (CaPTC) Reveals DNA Repair Genes Associated with African Ancestry.Cancer Res Commun. 2022 Sep 16;2(9):1005-1016. doi: 10.1158/2767-9764.CRC-22-0136. eCollection 2022 Sep. Cancer Res Commun. 2022. PMID: 36922933 Free PMC article.
-
Genomic analysis of benign prostatic hyperplasia implicates cellular re-landscaping in disease pathogenesis.JCI Insight. 2019 May 16;5(12):e129749. doi: 10.1172/jci.insight.129749. JCI Insight. 2019. PMID: 31094703 Free PMC article.
-
Spatial architectures of somatic mutations in normal prostate, benign prostatic hyperplasia and coexisting prostate cancer.Exp Mol Med. 2024 Feb;56(1):168-176. doi: 10.1038/s12276-023-01140-8. Epub 2024 Jan 4. Exp Mol Med. 2024. PMID: 38172600 Free PMC article.
-
Prostate stem cells in the development of benign prostate hyperplasia and prostate cancer: emerging role and concepts.Biomed Res Int. 2013;2013:107954. doi: 10.1155/2013/107954. Epub 2013 Jul 8. Biomed Res Int. 2013. PMID: 23936768 Free PMC article. Review.
-
Ancestry-Specific DNA Damage Repair Gene Mutations and Prostate Cancer.Cancers (Basel). 2025 Feb 18;17(4):682. doi: 10.3390/cancers17040682. Cancers (Basel). 2025. PMID: 40002276 Free PMC article. Review.
References
-
- Bushman W, Etiology, epidemiology, and natural history of benign prostatic hyperplasia. Urol Clin North Am, 2009. 36(4): p. 403–15, v. - PubMed
-
- Calogero AE, et al., Epidemiology and risk factors of lower urinary tract symptoms/benign prostatic hyperplasia and erectile dysfunction. Aging Male, 2019. 22(1): p. 12–19. - PubMed
-
- Berry SJ, et al., The development of human benign prostatic hyperplasia with age. J Urol, 1984. 132(3): p. 474–9. - PubMed
-
- Sarma AV, et al., Comparison of lower urinary tract symptom severity and associated bother between community-dwelling black and white men: the Olmsted County Study of Urinary Symptoms and Health Status and the Flint Men’s Health Study. Urology, 2003. 61(6): p. 1086–91. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

