Single-Cell Sequencing Reveals Functional Alterations in Tuberculosis
- PMID: 38192178
- PMCID: PMC10953544
- DOI: 10.1002/advs.202305592
Single-Cell Sequencing Reveals Functional Alterations in Tuberculosis
Abstract
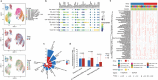
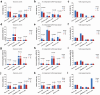
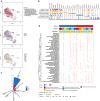
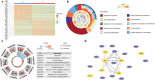
Despite its importance, the functional heterogeneity surrounding the dynamics of interactions between mycobacterium tuberculosis and human immune cells in determining host immune strength and tuberculosis (TB) outcomes, remains far from understood. This work now describes the development of a new technological platform to elucidate the immune function differences in individuals with TB, integrating single-cell RNA sequencing and cell surface antibody sequencing to provide both genomic and phenotypic information from the same samples. Single-cell analysis of 23 990 peripheral blood mononuclear cells from a new cohort of primary TB patients and healthy controls enables to not only show four distinct immune phenotypes (TB, myeloid, and natural killer (NK) cells), but also determine the dynamic changes in cell population abundance, gene expression, developmental trajectory, transcriptomic regulation, and cell-cell signaling. In doing so, TB-related changes in immune cell functions demonstrate that the immune response is mediated through host T cells, myeloid cells, and NK cells, with TB patients showing decreased naive, cytotoxicity, and memory functions of T cells, rather than their immunoregulatory function. The platform also has the potential to identify new targets for immunotherapeutic treatment strategies to restore T cells from dysfunctional or exhausted states.
Keywords: functional variation; heterogeneity; immunosuppression; single-cell sequencing; tuberculosis.
© 2024 The Authors. Advanced Science published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Bahuguna A., Rawat S., Rawat D. S., Med. Res. Rev. 2021, 41, 2565. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
