Extracellular vesicles released by cancer-associated fibroblast-induced myeloid-derived suppressor cells inhibit T-cell function
- PMID: 38192443
- PMCID: PMC10773711
- DOI: 10.1080/2162402X.2023.2300882
Extracellular vesicles released by cancer-associated fibroblast-induced myeloid-derived suppressor cells inhibit T-cell function
Abstract
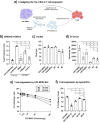
Myeloid cells are known to play a crucial role in creating a tumor-promoting and immune suppressive microenvironment. Our previous study demonstrated that primary human monocytes can be polarized into immunosuppressive myeloid-derived suppressor cells (MDSCs) by cancer-associated fibroblasts (CAFs) in a 3D co-culture system. However, the molecular mechanisms underlying the immunosuppressive function of MDSCs, especially CAF-induced MDSCs, remain poorly understood. Using mass spectrometry-based proteomics, we compared cell surface protein changes among monocytes, in vitro differentiated CAF-induced MDSCs, M1/M2 macrophages, and dendritic cells, and identified an extracellular vesicle (EV)-mediated secretory phenotype of MDSCs. Functional assays using an MDSC/T-cell co-culture system revealed that blocking EV generation in CAF-induced MDSCs reversed their ability to suppress T-cell proliferation, while EVs isolated from CAF-induced MDSCs directly inhibited T-cell function. Furthermore, we identified fructose bisphosphatase 1 (FBP1) as a cargo protein that is highly enriched in EVs isolated from CAF-induced MDSCs, and pharmacological inhibition of FBP1 partially reversed the suppressive phenotype of MDSCs. Our findings provide valuable insights into the cell surface proteome of different monocyte-derived myeloid subsets and uncover a novel mechanism underlying the interplay between CAFs and myeloid cells in shaping a tumor-permissive microenvironment.
Keywords: Cancer-associated fibroblasts (CAF); extracellular vesicles (EV); myeloid-derived suppressor cells (MDSC); proteomics.
© 2024 Merck & Co., Inc., Rahway, NJ, USA and its affiliates. Published with license by Taylor & Francis Group, LLC.
Conflict of interest statement
All authors are current or previous employees of Merck Sharp & Dohme LLC, a subsidiary of Merck & Co., Inc., Rahway, NJ, USA and shareholders of Merck & Co., Inc., Rahway, NJ, USA.
Figures



References
-
- Cristescu R, Nebozhyn M, Zhang C, Albright A, Kobie J, Huang L, Zhao Q, Wang A, Ma H, Alexander Cao Z, et al. Transcriptomic determinants of response to pembrolizumab monotherapy across solid tumor types. Clin Cancer Res. 2022;28(8):1680–1689. doi: 10.1158/1078-0432.CCR-21-3329. - DOI - PMC - PubMed
-
- Singh L, Muise ES, Bhattacharya A, Grein J, Javaid S, Stivers P, Zhang J, Qu Y, Joyce-Shaikh B, Loboda A, et al. ILT3 (LILRB4) promotes the immunosuppressive function of tumor-educated human monocytic myeloid-derived suppressor cells. Mol Cancer Res. 2021;19(4):702–716. doi: 10.1158/1541-7786.MCR-20-0622. - DOI - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
