Genetic analysis reveals a robust and hierarchical recruitment of the LolA chaperone to the LolCDE lipoprotein transporter
- PMID: 38193657
- PMCID: PMC10865981
- DOI: 10.1128/mbio.03039-23
Genetic analysis reveals a robust and hierarchical recruitment of the LolA chaperone to the LolCDE lipoprotein transporter
Abstract
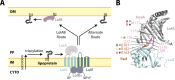
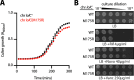
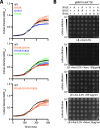
The outer membrane (OM) is an essential organelle of Gram-negative bacteria. Lipoproteins are key to building the OM, performing essential functions in several OM assembly machines. Lipoproteins mature in the inner membrane (IM) and are then trafficked to the OM. In Escherichia coli, the LolCDE transporter is needed to extract lipoproteins from the IM to begin trafficking. Lipoproteins are then transferred from LolCDE to the periplasmic chaperone LolA which ferries them to the OM for insertion by LolB. LolA recruitment by LolC is an essential trafficking step. Structural and biochemical studies suggested that two regions (termed Hook and Pad) within a periplasmic loop of LolC worked in tandem to recruit LolA, leading to a bipartite model for recruitment. Here, we genetically examine the LolC periplasmic loop in vivo using E. coli. Our findings challenge the bipartite interaction model. We show that while the Hook is essential for lipoprotein trafficking in vivo, lipoproteins are still efficiently trafficked when the Pad residues are inactivated. We show with AlphaFold2 multimer modeling that Hook:LolA interactions are likely universal among diverse Gram-negative bacteria. Conversely, Pad:LolA interactions vary across phyla. Our in vivo data redefine LolC:LolA recruitment into a hierarchical interaction model. We propose that the Hook is the major player in LolA recruitment, while the Pad plays an ancillary role that is important for efficiency but is ultimately dispensable. Our findings expand the understanding of a fundamental step in essential lipoprotein trafficking and have implications for efforts to develop new antibacterials that target LolCDE.IMPORTANCEResistance to current antibiotics is increasingly common. New antibiotics that target essential processes are needed to expand clinical options. For Gram-negative bacteria, their cell surface-the outer membrane (OM)-is an essential organelle and antibiotic barrier that is an attractive target for new antibacterials. Lipoproteins are key to building the OM. The LolCDE transporter is needed to supply the OM with lipoproteins and has been a focus of recent antibiotic discovery. In vitro evidence recently proposed a two-part interaction of LolC with LolA lipoprotein chaperone (which traffics lipoproteins to the OM) via "Hook" and "Pad" regions. We show that this model does not reflect lipoprotein trafficking in vivo. Only the Hook is essential for lipoprotein trafficking and is remarkably robust to mutational changes. The Pad is non-essential for lipoprotein trafficking but plays an ancillary role, contributing to trafficking efficiency. These insights inform ongoing efforts to drug LolCDE.
Keywords: LolA; LolC; LolCDE; lipoprotein; lipoprotein trafficking; outer membrane.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







Update of
-
Genetic analysis reveals a robust and hierarchical recruitment of the LolA chaperone to the LolCDE lipoprotein transporter.bioRxiv [Preprint]. 2023 Nov 8:2023.11.08.566237. doi: 10.1101/2023.11.08.566237. bioRxiv. 2023. Update in: mBio. 2024 Feb 14;15(2):e0303923. doi: 10.1128/mbio.03039-23. PMID: 37986794 Free PMC article. Updated. Preprint.
Similar articles
-
Genetic analysis reveals a robust and hierarchical recruitment of the LolA chaperone to the LolCDE lipoprotein transporter.bioRxiv [Preprint]. 2023 Nov 8:2023.11.08.566237. doi: 10.1101/2023.11.08.566237. bioRxiv. 2023. Update in: mBio. 2024 Feb 14;15(2):e0303923. doi: 10.1128/mbio.03039-23. PMID: 37986794 Free PMC article. Updated. Preprint.
-
Structural basis of lipoprotein recognition by the bacterial Lol trafficking chaperone LolA.Proc Natl Acad Sci U S A. 2022 Sep 6;119(36):e2208662119. doi: 10.1073/pnas.2208662119. Epub 2022 Aug 29. Proc Natl Acad Sci U S A. 2022. PMID: 36037338 Free PMC article.
-
Teasing apart the evolution of lipoprotein trafficking in gram-negative bacteria reveals a bifunctional LolA.Proc Natl Acad Sci U S A. 2023 Feb 7;120(6):e2218473120. doi: 10.1073/pnas.2218473120. Epub 2023 Jan 30. Proc Natl Acad Sci U S A. 2023. PMID: 36716372 Free PMC article.
-
Lipoproteins and Their Trafficking to the Outer Membrane.EcoSal Plus. 2019 Mar;8(2):10.1128/ecosalplus.ESP-0038-2018. doi: 10.1128/ecosalplus.ESP-0038-2018. EcoSal Plus. 2019. PMID: 30900542 Free PMC article. Review.
-
Sorting of lipoproteins to the outer membrane in E. coli.Biochim Biophys Acta. 2004 Nov 11;1694(1-3):IN1-9. Biochim Biophys Acta. 2004. PMID: 15672528 Review.
Cited by
-
LPS O-antigen polysaccharide length impacts outer membrane permeability of enteric gram-negative bacteria.bioRxiv [Preprint]. 2025 Aug 15:2025.08.14.670410. doi: 10.1101/2025.08.14.670410. bioRxiv. 2025. PMID: 40832261 Free PMC article. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
