Escherichia coli sequence type 410 with carbapenemases: a paradigm shift within E. coli toward multidrug resistance
- PMID: 38193668
- PMCID: PMC10869336
- DOI: 10.1128/aac.01339-23
Escherichia coli sequence type 410 with carbapenemases: a paradigm shift within E. coli toward multidrug resistance
Abstract
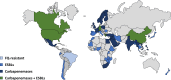
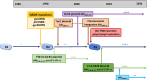
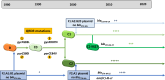
Escherichia coli sequence type ST410 is an emerging carbapenemase-producing multidrug-resistant (MDR) high-risk One-Health clone with the potential to significantly increase carbapenem resistance among E. coli. ST410 belongs to two clades (ST410-A and ST410-B) and three subclades (ST410-B1, ST410-B2, and ST410-B3). After a fimH switch between clades ST410-A and ST410-B1, ST410-B2 and ST410-B3 subclades showed a stepwise progression toward developing MDR. (i) ST410-B2 initially acquired fluoroquinolone resistance (via homologous recombination) in the 1980s. (ii) ST410-B2 then obtained CMY-2, CTX-M-15, and OXA-181 genes on different plasmid platforms during the 1990s. (iii) This was followed by the chromosomal integration of blaCMY-2, fstl YRIN insertion, and ompC/ompF mutations during the 2000s to create the ST410-B3 subclade. (iv) An IncF plasmid "replacement" scenario happened when ST410-B2 transformed into ST410-B3: F36:31:A4:B1 plasmids were replaced by F1:A1:B49 plasmids (both containing blaCTX-M-15) followed by blaNDM-5 incorporation during the 2010s. User-friendly cost-effective methods for the rapid identification of ST410 isolates and clades are needed because limited data are available about the frequencies and global distribution of ST410 clades. Basic mechanistic, evolutionary, surveillance, and clinical studies are urgently required to investigate the success of ST410 (including the ability to acquire successive MDR determinants). Such information will aid with management and prevention strategies to curb the spread of carbapenem-resistant E. coli. The medical community can ill afford to ignore the spread of a global E. coli clone with the potential to end the carbapenem era.
Keywords: Escherichia coli; ST410; carbapenemases; high-risk clones.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Acquisition of genomic elements were pivotal for the success of Escherichia coli ST410.J Antimicrob Chemother. 2022 Nov 28;77(12):3399-3407. doi: 10.1093/jac/dkac329. J Antimicrob Chemother. 2022. PMID: 36204996 Free PMC article.
-
Dissemination of an international high-risk clone of Escherichia coli ST410 co-producing NDM-5 and OXA-181 carbapenemases in Seoul, Republic of Korea.Int J Antimicrob Agents. 2021 Dec;58(6):106448. doi: 10.1016/j.ijantimicag.2021.106448. Epub 2021 Oct 11. Int J Antimicrob Agents. 2021. PMID: 34648943
-
Escherichia coli Sequence Type 410 Is Causing New International High-Risk Clones.mSphere. 2018 Jul 18;3(4):e00337-18. doi: 10.1128/mSphere.00337-18. mSphere. 2018. PMID: 30021879 Free PMC article.
-
Escherichia coli ST1193: Following in the Footsteps of E. coli ST131.Antimicrob Agents Chemother. 2022 Jul 19;66(7):e0051122. doi: 10.1128/aac.00511-22. Epub 2022 Jun 6. Antimicrob Agents Chemother. 2022. PMID: 35658504 Free PMC article. Review.
-
The Significance of Epidemic Plasmids in the Success of Multidrug-Resistant Drug Pandemic Extraintestinal Pathogenic Escherichia coli.Infect Dis Ther. 2023 Apr;12(4):1029-1041. doi: 10.1007/s40121-023-00791-4. Epub 2023 Mar 22. Infect Dis Ther. 2023. PMID: 36947392 Free PMC article. Review.
Cited by
-
Mobile genetic elements of global Escherichia coli ST131 clades with carbapenemases.Eur J Clin Microbiol Infect Dis. 2025 Jun 7. doi: 10.1007/s10096-025-05187-5. Online ahead of print. Eur J Clin Microbiol Infect Dis. 2025. PMID: 40481333
-
A new in vivo model of intestinal colonization using Zophobas morio larvae: testing hyperepidemic ESBL- and carbapenemase-producing Escherichia coli clones.Front Microbiol. 2024 Apr 10;15:1381051. doi: 10.3389/fmicb.2024.1381051. eCollection 2024. Front Microbiol. 2024. PMID: 38659985 Free PMC article.
-
Genomic Epidemiology of Pseudomonas aeruginosa Sequence Type 111.Eur J Clin Microbiol Infect Dis. 2025 Feb;44(2):375-381. doi: 10.1007/s10096-024-05010-7. Epub 2024 Dec 11. Eur J Clin Microbiol Infect Dis. 2025. PMID: 39658728
-
Derivatization of pBACpAK entrapment vectors for enhanced mobile genetic element transposition detection in multidrug-resistant Escherichia coli.Access Microbiol. 2025 May 23;7(5):001013.v3. doi: 10.1099/acmi.0.001013.v3. eCollection 2025. Access Microbiol. 2025. PMID: 40416558 Free PMC article.
-
Successful management of a multi-species outbreak of carbapenem-resistant organisms in Fiji: a prospective genomics-enhanced investigation and response.Lancet Reg Health West Pac. 2024 Nov 20;53:101234. doi: 10.1016/j.lanwpc.2024.101234. eCollection 2024 Dec. Lancet Reg Health West Pac. 2024. PMID: 39633714 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

