The structure and assembly mechanisms of T4-like cyanophages community in the South China Sea
- PMID: 38193726
- PMCID: PMC10846272
- DOI: 10.1128/spectrum.02002-23
The structure and assembly mechanisms of T4-like cyanophages community in the South China Sea
Abstract
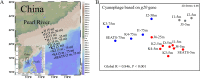
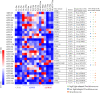
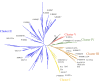
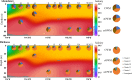
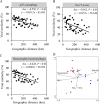
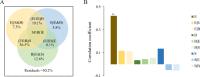
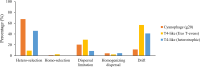
Marine ecosystems contain an immense diversity of phages, many of which infect cyanobacteria (cyanophage) that are largely responsible for primary productivity. To characterize the genetic diversity and biogeographic distribution of the marine T4-like cyanophage community in the northern South China Sea, the T4-like cyanophage portal protein gene (g20) was amplified. Phylogenetic analysis revealed that marine T4-like cyanophages were highly diverse, with g20 operational taxonomic units being affiliated with five defined clades (Clusters I-V). Cluster II had a wide geographic distribution, Cluster IV was the most abundant in the open sea, and Cluster I was dominant in coastal shelf environments. Our results showed T4-like cyanophages (based on g20) community was generally shaped via heterogeneous selection. Highly variable environmental factors (such as salinity and temperature) can heterogeneously select different cyanophage communities. Nevertheless, the dominant drivers of the T4-like cyanophage community based on the g20 and g23 (T4-like phage major capsid protein gene) were different, probably due to different coverages by the primer sets. Furthermore, the community assembly processes of T4-like cyanophages were affected by host traits (abundance and distribution), viral traits (latent period, burst size, and host range), and environmental properties (temperature and salinity).IMPORTANCECyanophages are abundant and ubiquitous in the oceans, altering population structures and evolution of cyanobacteria, which account for a large portion of global carbon fixation, through host mortality, horizontal gene transfer, and the modulation of host metabolism. However, little is known about the biogeography and ecological drivers that shape the cyanophage community. Here, we use g20 and g23 genes to examine the biogeographic patterns and the assembly mechanisms of T4-like cyanophage community in the northern part of the South China Sea. The different coverages of primer sets might lead to the different dominant drivers of T4-like cyanophage community based on g20 and g23 genes. Our results showed that characteristics of viral traits (latent period, burst size, and host range) and host traits (abundance and distribution) were found to either limit or enhance the biogeographic distribution of T4-like cyanophages. Overall, both virus and host properties are critical to consider when determining rules of community assembly for viruses.
Keywords: South China Sea; T4-like cyanophage; community assembly; g20.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Phylogenetic diversity of marine cyanophage isolates and natural virus communities as revealed by sequences of viral capsid assembly protein gene g20.Appl Environ Microbiol. 2002 Apr;68(4):1576-84. doi: 10.1128/AEM.68.4.1576-1584.2002. Appl Environ Microbiol. 2002. PMID: 11916671 Free PMC article.
-
A Unique Set of Auxiliary Metabolic Genes Found in an Isolated Cyanophage Sheds New Light on Marine Phage-Host Interactions.Microbiol Spectr. 2022 Oct 26;10(5):e0236722. doi: 10.1128/spectrum.02367-22. Epub 2022 Oct 3. Microbiol Spectr. 2022. PMID: 36190421 Free PMC article.
-
Phylogenetic distribution of the capsid assembly protein gene (g20) of cyanophages in paddy floodwaters in Northeast China.PLoS One. 2014 Feb 12;9(2):e88634. doi: 10.1371/journal.pone.0088634. eCollection 2014. PLoS One. 2014. PMID: 24533125 Free PMC article.
-
[Genetic diversity of capsid assembly protein genes (g20) of cyanophage in different natural environment--a review].Wei Sheng Wu Xue Bao. 2013 Nov 4;53(11):1149-57. Wei Sheng Wu Xue Bao. 2013. PMID: 24617255 Review. Chinese.
-
From natural to artificial cyanophages: Current progress and application prospects.Environ Res. 2023 Apr 15;223:115428. doi: 10.1016/j.envres.2023.115428. Epub 2023 Feb 4. Environ Res. 2023. PMID: 36746205 Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

