Genomic Classification and Individualized Prognosis in Multiple Myeloma
- PMID: 38194610
- PMCID: PMC11095887
- DOI: 10.1200/JCO.23.01277
Genomic Classification and Individualized Prognosis in Multiple Myeloma
Abstract
Purpose: Outcomes for patients with newly diagnosed multiple myeloma (NDMM) are heterogenous, with overall survival (OS) ranging from months to over 10 years.
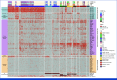
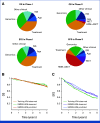
Methods: To decipher and predict the molecular and clinical heterogeneity of NDMM, we assembled a series of 1,933 patients with available clinical, genomic, and therapeutic data.
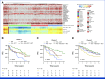
Results: Leveraging a comprehensive catalog of genomic drivers, we identified 12 groups, expanding on previous gene expression-based molecular classifications. To build a model predicting individualized risk in NDMM (IRMMa), we integrated clinical, genomic, and treatment variables. To correct for time-dependent variables, including high-dose melphalan followed by autologous stem-cell transplantation (HDM-ASCT), and maintenance therapy, a multi-state model was designed. The IRMMa model accuracy was significantly higher than all comparator prognostic models, with a c-index for OS of 0.726, compared with International Staging System (ISS; 0.61), revised-ISS (0.572), and R2-ISS (0.625). Integral to model accuracy was 20 genomic features, including 1q21 gain/amp, del 1p, TP53 loss, NSD2 translocations, APOBEC mutational signatures, and copy-number signatures (reflecting the complex structural variant chromothripsis). IRMMa accuracy and superiority compared with other prognostic models were validated on 256 patients enrolled in the GMMG-HD6 (ClinicalTrials.gov identifier: NCT02495922) clinical trial. Individualized patient risks were significantly affected across the 12 genomic groups by different treatment strategies (ie, treatment variance), which was used to identify patients for whom HDM-ASCT is particularly effective versus patients for whom the impact is limited.
Conclusion: Integrating clinical, demographic, genomic, and therapeutic data, to our knowledge, we have developed the first individualized risk-prediction model enabling personally tailored therapeutic decisions for patients with NDMM.
Conflict of interest statement
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (
Figures



References
-
- D'Agostino M, Cairns DA, Lahuerta JJ, et al. Second revision of the International Staging System (R2-ISS) for overall survival in multiple myeloma: A European Myeloma Network (EMN) report within the HARMONY project. J Clin Oncol. 2022;40:3406–3418. - PubMed
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

